•
Author: Rosa Fernández García •
∞
Note: We are rebooting our “Life in the Lab” series, which features interviews of interns and visitors. This post is by our inaugural OMA Visiting Fellow Rosa Fernández García, who spent a month with us earlier this year. You can follow Rosa on Twitter at @Rosamygale. —Christophe
Please introduce yourself and your research interests.
I received my bachelor’s degree in Biology (major in Zoology) at Complutense University in Madrid, Spain. I got my master’s and PhD at the same university with a thesis about phylogeny and phylogeography of cosmopolitan earthworms. After that, I moved to the lab of Prof. Gonzalo Giribet at Harvard University where I was a postdoc during 3 years and a Research Associate for another year. In January 2017, I’ll move to Barcelona to work as a Research Fellow in the lab of Dr. Toni Gabaldón at the Center for Genomic Regulation.
My research addresses fundamental questions about evolution in invertebrates: in other words, I am fascinated by how, when and where biodiversity took its form, and why it is maintained. My main two animal groups of interest are terrestrial annelids (oligochaetes) and (pan)arthropods, particularly the earliest branching lineages and most scientifically neglected groups (chelicerates and myriapods).
How did biodiversity took its shape? Resolving the tree of life. Macroevolutionary patterns are generally what we see when we look at the large-scale history of life. It encompasses the grandest trends and transformations in evolution, such as the origin of bilateral animals or the radiation of arthropods. In order to understand how lineages are related to each other, I study macroevolutionary patterns in several groups of invertebrates through phylogenetics and phylogenomics. I currently lead a fruitful line of research dealing with phylogenomics of myriapods and chelicerates, having optimized protocols to sequence successfully single individuals of the rarest and smallest arthropods. We are getting closer to resolve the Arthropod Tree of Life!
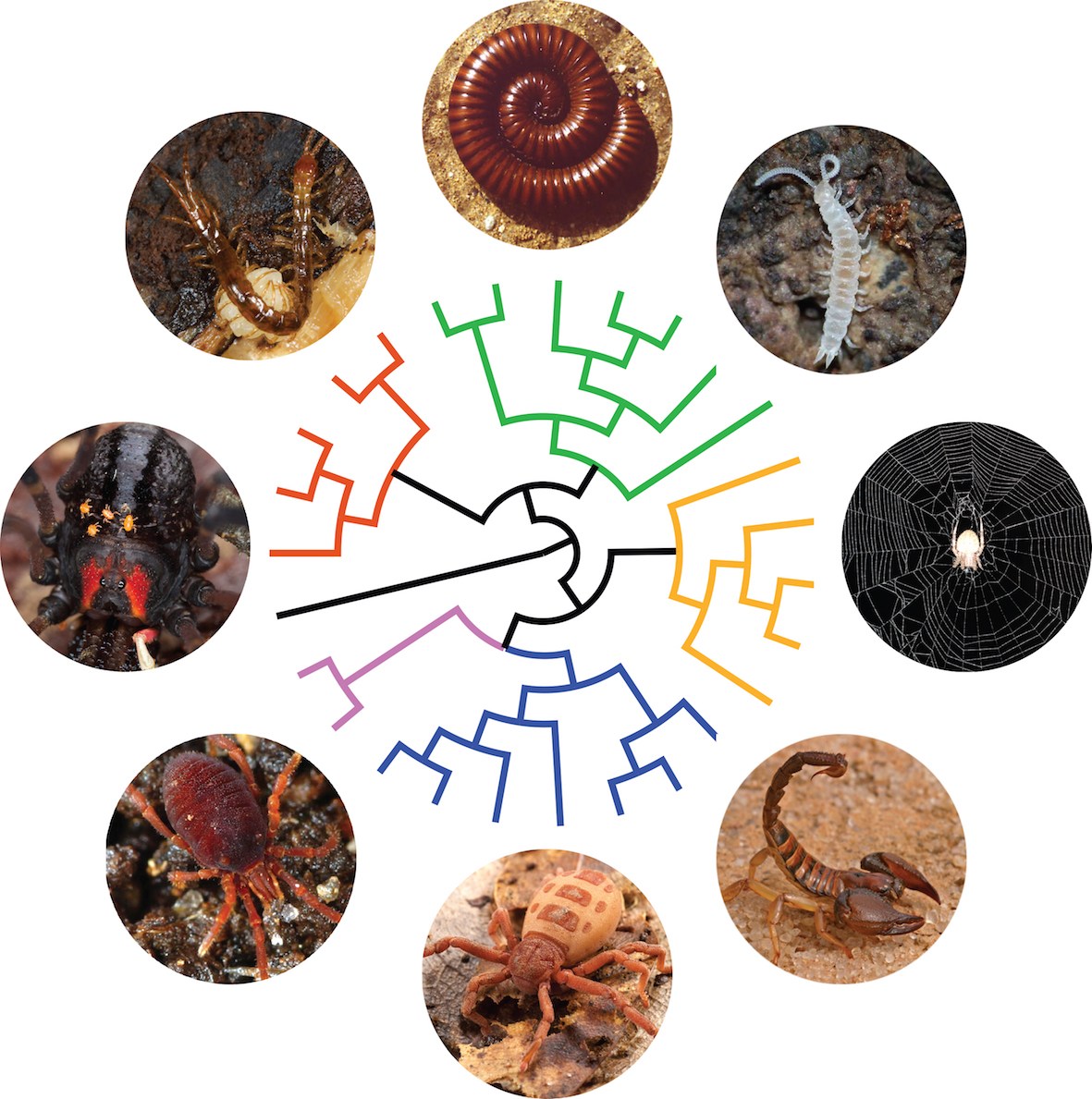
Artist’s rendition of the Arthropod tree of life
When and where? I tried to understand the mode and tempo of animal diversification patterns through the integration of phylogeography, biogeography and paleogeography.
Why? Comparative transcriptomics and genomics is a very powerful tool to shed light on very interesting evolutionary questions, such as arthropod terrestrialization - one of my favorite new lines of research.
Why did you choose to apply to the OMA visiting fellowship programme?
Orthology inference is one of the key steps in phylogenomics. I had been using OMA for a few years and I wanted to learn how I could use it more efficiently in my ongoing projects.
What project did you work on during your visit?
My project focused on optimizing OMA runs for some big and challenging data sets that I was having problems with. Also, I was interested in learning how I could exploit hierarchical orthogroups for comparative genomics studies in arthropods.

Rosa and David Dylus at coffee break (photo by Arthur Dessimoz)
Was there any highlight or low point you’d like to share?
It was a great experience to be in the Dessimoz lab for a month. As a systematist with relatively limited bioinformatic background, it was absolutely great to exchange ideas with computer scientists interested in the same scientific problems but with a completely different perspective that mine. It was a very enriching experience.
Do you have any practical tip for future OMA visiting fellows?
One month was not enough for me, so try to stay longer if your project is ambitious. And ask Christophe to bring a Tête de Moine cheese in your last day, it’s delicious!
Editor’s note: If you are interested in the OMA visiting fellowship programme, consult this page.
References:
Fernández R, Laumer CE, Vahtera V, Libro S, Kaluziak S, Sharma PP, Pérez-Porro AR, Edgecombe GD, & Giribet G (2014). Evaluating topological conflict in centipede phylogeny using transcriptomic data sets. Molecular biology and evolution, 31 (6), 1500-13 PMID: 24674821
Fernández, R., Hormiga, G., & Giribet, G. (2014). Phylogenomic Analysis of Spiders Reveals Nonmonophyly of Orb Weavers Current Biology, 24 (15), 1772-1777 DOI: 10.1016/j.cub.2014.06.035
Fernández R, & Giribet G (2015). Unnoticed in the tropics: phylogenomic resolution of the poorly known arachnid order Ricinulei (Arachnida). Royal Society open science, 2 (6) PMID: 26543583
Novo M, Fernández R, Andrade SC, Marchán DF, Cunha L, & Díaz Cosín DJ (2016). Phylogenomic analyses of a Mediterranean earthworm family (Annelida: Hormogastridae). Molecular phylogenetics and evolution, 94 (Pt B), 473-8 PMID: 26522608
Fernández R, Edgecombe GD, & Giribet G (2016). Exploring Phylogenetic Relationships within Myriapoda and the Effects of Matrix Composition and Occupancy on Phylogenomic Reconstruction. Systematic biology, 65 (5), 871-89 PMID: 27162151
Sharma, P., Fernandez, R., Esposito, L., Gonzalez-Santillan, E., & Monod, L. (2015). Phylogenomic resolution of scorpions reveals multilevel discordance with morphological phylogenetic signal Proceedings of the Royal Society B: Biological Sciences, 282 (1804), 20142953-20142953 DOI: 10.1098/rspb.2014.2953
Rosa Fernandez, Prashant Sharma, Ana LM Tourinho, & Gonzalo Giribet (2016). The Opiliones Tree of Life: shedding light on harvestmen relationships through transcriptomics BioRxiv DOI: 10.1101/077594
•
Author: Tunca Doğan •
∞
Note: the “Life in the Lab” series features interviews of interns and visitors. This post is by our second 2016 OMA Visiting Fellow Tunca Doğan, who spent a month with us earlier this year. You can follow Tunca on Twitter at @tuncadogan. —Christophe
Please introduce yourself in a few sentences.
My name is Tunca Doğan. I received my PhD in 2013 with a thesis study in the fields of bioinformatics and computational biology where we developed methods for the clustering of the protein sequences using unsupervised machine learning techniques (Dogan and Karacali, 2013). I’ve since been working as a post-doctoral fellow in the EMBL-EBI, UK under the Protein Function Development team (UniProt Database) leaded by Dr Maria Martin. Here I’m developing new tools and methods for the automated functional annotation of protein records in the UniProtKB using a variety of features including domain architectures (Dogan et al., 2016). I’m also conducting research in the field of computational drug discovery. As of 2016, I’m also affiliated to the Department of Health Informatics, METU, Turkey both as a senior research fellow and a faculty candidate.
Why did you choose to apply to the OMA visiting fellowship programme?
The team behind OMA is world-leading in the field of phylogenomics, and they authored many highly cited publications in this area. Moreover, OMA is considered to be one of the most reliable and comprehensive resources offering phylogenomic information on various species. I’ve applied to this programme in order to develop my knowledge in phylogenomic research, particularly about the OMA production. My specific research aim was to investigate if and how the information in OMA can be utilized in order to increase the coverage and the quality of the automated functional annotation of proteins in the UniProt database.

Discussions on UNIL campus with Leonardo de Oliveria Martins, Surag Nair, Clément Train, David Dylus and Tunca Doğan (from l. to r.)
What project did you work on during your visit?
The project I worked on had two sides: 1) investigating novel ways of quality checking of the data produced in the OMA pipeline (especially HOGs) using the Domain Architecture Alignment and Classification (DAAC) method I previously developed in UniProt; 2) investigating the use of OMA groups and HOGs to propagate the functional annotation between the (homologous) member proteins of the same clusters/classes.
Was there any highlight or low point you’d like to share?
It was a great experience for me both professionally and socially. I’ve learnt a great deal in just one month and we still keep our collaboration with the continuation of the abovementioned project. Everyone I met in the group: Christophe, Adrian, David, Leonardo and Clement were all knowledgeable, helpful and friendly that I had great time during my stay. It was a great pleasure to meet and to work with them all…
UNIL/EPFL campus is just beautiful, at the shores of lake Geneva. The campus is also well-equipped for all possible needs. This was also my first time in Switzerland and I was enchanted by the beauty of this country… The only downside for a foreign visitor could be the expensiveness of life in Switzerland, which was also manageable with a little prior investigation and planning.
Do you have any practical tip for future OMA visiting fellows?
I definitely recommend any researcher (at PhD or post-doc level) that has an interest in phylogenomics to apply to this programme. You’ll learn a great deal and have a good time at the same time. Also (for the foreigners) do not forget about travelling around this beautiful country in your spare time…
Editor’s note: If you are interested in the OMA visiting fellowship programme, consult this page.
References:
Doğan T, & Karaçalı B (2013). Automatic identification of highly conserved family regions and relationships in genome wide datasets including remote protein sequences. PloS one, 8 (9) PMID: 24069417
Doğan T, MacDougall A, Saidi R, Poggioli D, Bateman A, O’Donovan C, & Martin MJ (2016). UniProt-DAAC: domain architecture alignment and classification, a new method for automatic functional annotation in UniProtKB. Bioinformatics (Oxford, England), 32 (15), 2264-71 PMID: 27153729