•
Author: Kevin Gori •
∞
This is how molecular systematics has worked since the sixties: you take some identifiable feature (e.g. a gene or a protein) common to a group of species and take some measurements of it (e.g. sequencing the DNA). By comparing the results of these measurements you can estimate the evolutionary tree that links the species. Shortly after people started doing this they realised there was a problem: when analyses are based different genes they often estimate different —incongruent — evolutionary trees. As technology has become more capable researchers have begun using more and more genes, so this problem of incongruent trees has moved to the foreground.
There have been lots of good ideas of what do about this problem, and this paper is our contribution. We tried to tackle incongruence by designing a method that groups genes together based on how similar their estimated trees are, without any assumption as to how any incongruence came about.
If all the genes more or less agree on the evolutionary tree, then you get one large group; if some disagree, then they are placed in their own groups. The most interesting case is if several genes disagree in the same way, because then you have an effect to try to explain, and you may have discovered something.
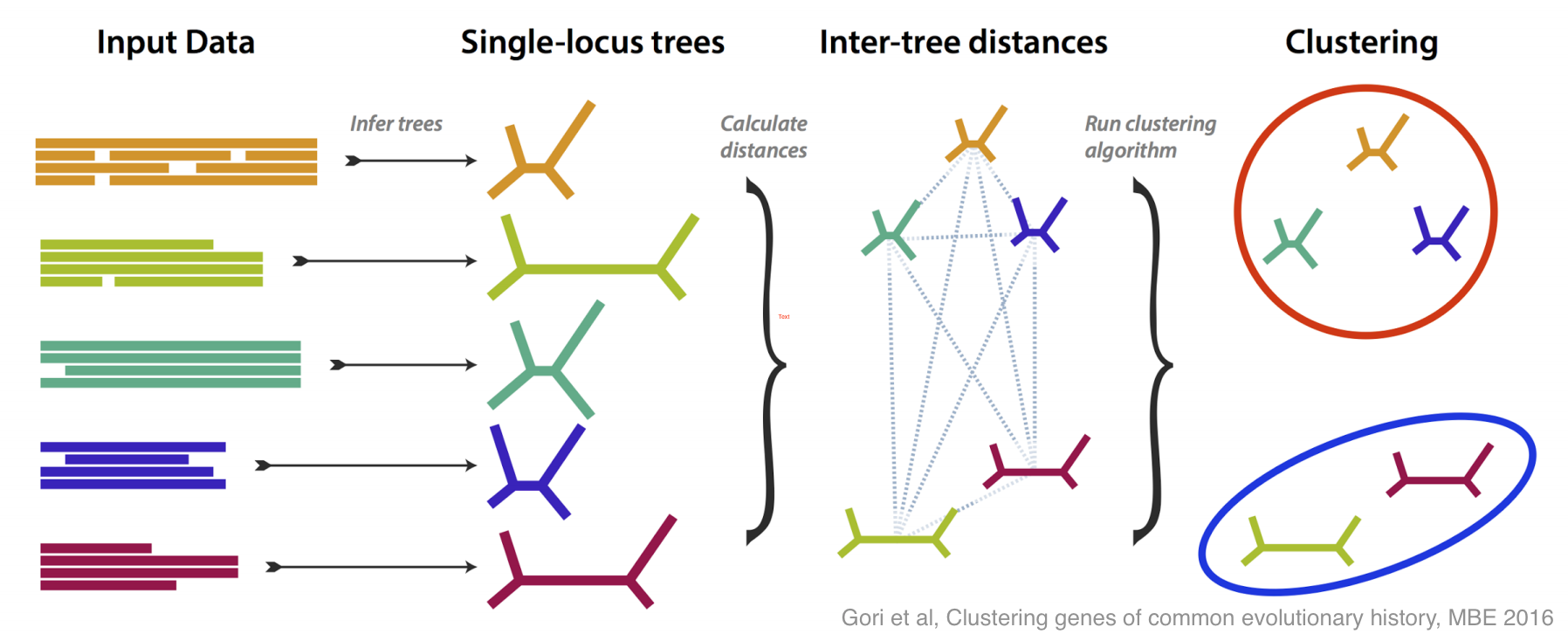
We did lots of simulation to test and refine our method, both in its ability to recognise different incongruent groups, and to estimate how many groups are present. Then, armed with a method that works well on simulation, we tested it on some real data, from yeasts, and from flies.
Our findings were that for the yeast data our method worked really well, and identified 3 distinct groups of genes. The majority of genes were a good fit to the widely accepted tree for the species we looked at. The other two groups showed some major differences, mostly involving two of the species. We had a close look at the data, and concluded that there were some wrong annotations in the data that had introduced sequences that didn’t belong there. This was not the biological result we were looking for, but nonetheless useful.
The flies data were more tricky, as they come from a genus where we aren’t sure how many separate species there are. We produced trees that show better species level resolution than the most recent molecular studies. We also showed high levels of incongruence in the order that the species appear, which can often be the case when species have diverged rapidly, due to a process called incomplete lineage sorting.
So be it to identify artifacts or genuine incongruence among your loci, we think that process-agnostic topology partitioning should become a routine step in phylogenetic analyses. To facilitate this process, we’ve released our code in a new open source software called “treeCl”, available at https://git.io/treeCl.
Reference
Gori K, Suchan T, Alvarez N, Goldman N, & Dessimoz C (2016). Clustering genes of common evolutionary history. Molecular biology and evolution PMID: 26893301
•
Author: Natasha Glover •
∞
We know homologs are genes related by common ancestry. But throw complex evolutionary events into the mix and things can get little dicey. Under the umbrella of homologs exist many different categories: orthologs, paralogs, ohnologs, xenologs, co-ortholog, in-paralogs, out-paralogs, paleologs, among others. All of these —log terms have a specific meaning (see my previous blog post on orthology and paralogy), but now we will focus on one in particular: homoeologs.
But before we get into the definition, let’s start at the beginning. When I started as a postdoc at Bayer CropScience working with Henning Redestig in collaboration with Christophe Dessimoz University College London, I was tasked with evaluating homoeolog predictions using the OMA algorithm.
What are homoeologs?
From my previous experience, I knew homoeologs as roughly “corresponding” genes between subgenomes of a polyploid organism. For example, the wheat genome is an allohexaploid, with 3 diploid subgenomes named A, B, and D. Given a gene on chromosome 3B, you will most likely find a nearly identical copy on chromosomes 3A and 3D, in roughly the same position. These corresponding copies across subgenomes are known as homoeologs. But this definition left something to be desired— it didn’t tell me anything about the evolutionary relationship between the homoeologs. Worse, it was ambiguous in that it required discretionary similarity thresholds in terms of sequence and positional conservation. How could we test for performance if there was no unambiguous definition of the target?
Time to hit the books
Like many researchers starting a new project, I went to the scientific literature to get more information. After many hours spent on google scholar, I found myself with more questions than answers. Firstly, what were the evolutionary events that give rise to homoeologs? How do they fit in with the other —log terms? Can they be found only in a certain type of polyploid, but not another? How do things like gene duplication and movement affect our understanding of what a homoeolog is? And finally, after seeing it the word written as homoeolog, homeolog, and homoeologue, how do you even spell it?
There are some excellent review papers out there on polyploidy which shed light on the biological consequences of homoeology. This, this, or this for example. However, when searching the whole of the literature, I found many inconsistent, vague, or even incorrect usages of the term homoeolog. Sometimes people defined homoeologs on the basis of their chromosome pairing patterns. Other times homoeologs were used to describe corresponding genes from different, although closely related species. Many papers said homoeologs were necessarily syntenic. Others don’t define the term at all.
Getting on the same page
These imprecise or incorrect definitions can lead to confusion. In recent years, advances in technology has afforded us the opportunity to sequence many new genomes, including polyploids. All these new techniques and have exploded the amount of data and brought about collaborations between geneticists, molecular biologists, plant breeders, bioinformaticians, phylogeneticists, and statisticians. Therefore we think it’s important to have a precise and evolutionary meaningful definition of homoeology as a reference point.
What we learned
Thus we went back to the earliest usage of the term we could find and synthesizing the literature to date. We define homoeologs as “pairs of genes or chromosomes in the same species that originated by speciation and were brought back together in the same genome by allopolyploidization”. For recent hybrids, as long as there was no rearrangement across subgenomes, homoeologs can be thought of as orthologs between these subgenomes. Here’s how they fit in with other common homologs:
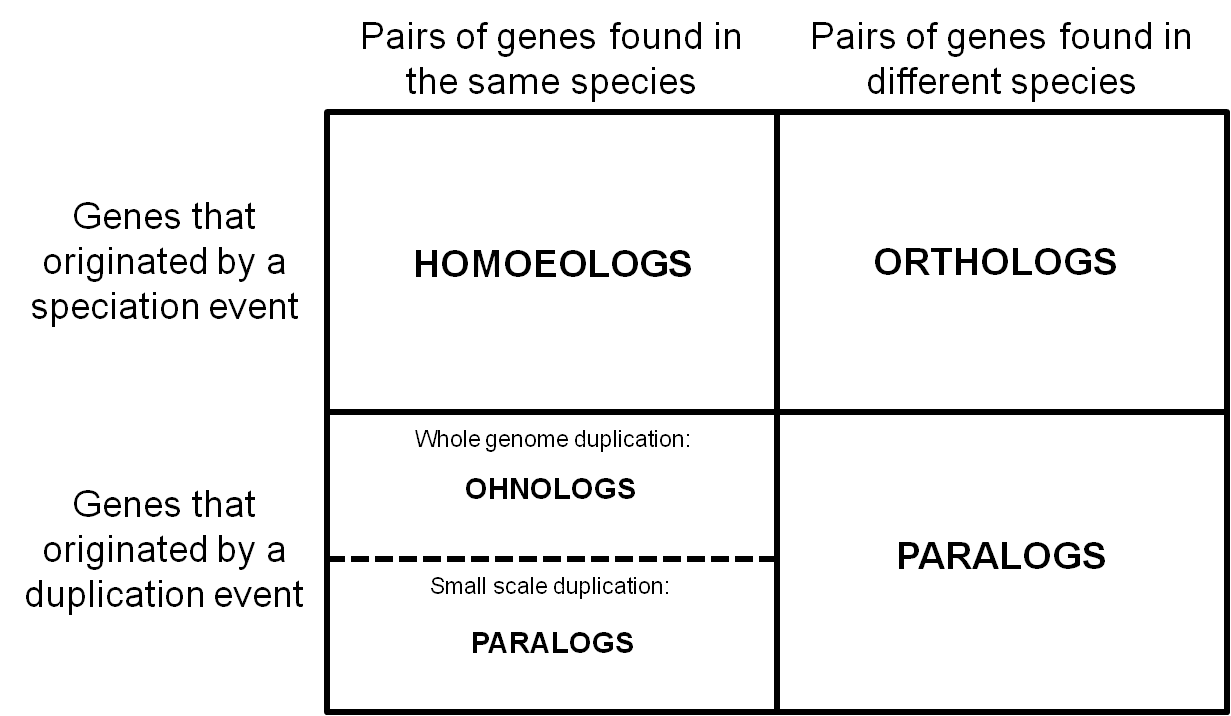
We realized that homoeologs are not necessarily one-to-one or syntenic. Depending on the particular patterns of gene duplication and rearrangement in a given species, we may see homoeologs at a 1:many or across non-corresponding chromosomes.
We also reviewed homoeolog inference techniques, starting from low-throughput lab techniques to evolution-based computational methods. Orthology prediction is a booming area of active research, so many orthology inference methods can be applied to homoeology prediction.
Last but not least, we learned that even though homoeolog has alternatively been spelled “homeolog” (no extra o), homoeolog is the clear winner in terms of popularity. The “homoeo—” spelling has been used more than double the amount of times in the literature. Fortunately however, both are pronounced the same (“ho-mee-o-log”)
Check out the review paper in Trends in Plant Science (open access!). We hope this paper can serve as a jump off point for those interested in tackling homoeology, especially for those new to the field.
Reference
Glover, N., Redestig, H., & Dessimoz, C. (2016). Homoeologs: What Are They and How Do We Infer Them? Trends in Plant Science DOI: 10.1016/j.tplants.2016.02.005