•
Author: Christophe Dessimoz •
∞
We are delighted to host Dr. Fritz Sedlazeck, from Prof. Arndt von Haeseler’s lab, for a special seminar:
- Title: The impact of highly polymorphic regions on high throughput sequencing related studies.
- Speaker: Dr. Fritz Sedlazeck
- Date, time & location: Thu 25 Sep 2014, 12 noon, Darwin Building Room 114
- Host: Christophe Dessimoz
- Abstract: The advent of high throughput sequencing (HTS) has boosted the variety of sequencing projects related to molecular biology and medicine. Mapping reads to a reference genome is one of the fundamental steps in most HTS related analysis. We show that highly polymorphic regions hinder an accurate alignment of HTS reads. Thus biasing subsequent analyses (e.g. SNP detection and transcription abundance estimation). We studied effect of such bias, by identifying highly polymorphic genomic regions in an F1 cross from inbred lines of D.m Mel 6 x D. m RAL774. To this end we determined all heterozygous positions in F1 from a genomic alignment. Each such heterozygous SNP was then categorized according to the number of SNPs in its vicinity. Mapping the RNA-seq data of the F1 cross to Mel 6D showed that neither BWA nor Tophat2 could recover most of the heterozygous SNPs alignment, where the detection probability depended on the variability around the SNP. Moreover, the heterozygous SNPs show an overrepresentation of the Mel 6D variant, thus the SNP frequencies deviate substantially from the expected value 0.5 per site. Furthermore, we demonstrate that a highly polymorphic region in a gene influences the estimation of its transcript abundance. To conclude, in this study, we demonstrate some read mappers are affected by highly polymorphic. We also show that mappers like NextGenMap are less affected and thus more suitable for reliable analyses of HTS data.
All welcome!
•
Author: Clément Train •
∞
Note: Inspired by the blog of Marcel Salathé’s group at Penn State, I have asked our recent interns to provide a brief account of their experience in the lab. They are posted in this new series “Life in the Lab”, unedited —Christophe
Please introduce yourself in a few sentences
I’m Clement Train and I’am a French student in bioinformatician at the University of Bordeaux. I studied biology but at some point I decided to learn computer science to try to solve biological problems. I’m passionate about evolution, genetics and all the fields that help to understand how life is created, regulated or can adapt to different environment.
Why did you choose to join the lab?
I was looking for an internship in bioinformatics for my 2014 summer. When I discovered the Dessimoz lab works on topic I am really interested in (molecular evolution, genetic, bioinformatics) so I decided to seize the opportunity to apply and Christophe let me join his lab as an intern.
What project did you work on (and for how long)?
I joined the team from June to August 2014. At first I worked on a web widget to visualize and select genomes in a phylogenetic tree. This project consisted in improving users’ utilization on the export function of the OMA browser for the OMA standalone. Afterward I worked on a face-lifting of the OMA browser website: we built a new structure for the website where users can easily learn what is OMA and how they can work with it. Plus we added new features like the synteny viewer.

Face-lifting of the OMA browser website
What came out of it?
This internship provided me with a great working experience as a bioinformatician. I learnt how to work with lots of people on different projects and how to adapt to new situations.
Was there any highlight or low point you’d like to share?
First i want to say that the mentorship was a very good motivation for me. All the team was following my project by helping me or giving me feedbacks to improve it. I have never felt alone in front of my computer and feeling that people were interested in my work was something I really enjoyed during my internship: to work on something where people are interest of. Second, I’d say that working on something that other people can use or reuse for them own work is very rewarding. Finally, I really liked the team atmosphere, lab retreat and lunch with everyone helped me feel comfortable during my whole internship and allowed me to create strong links with others team mates.
What is your overall impression and would you do it again?
I am completely happy with this internship. I met lots of good and funny people to work with and it is very nice to feel comfortable at work and can enjoy time with the team. I worked on great project where I was able to totally express myself because Christophe and Adrian had confidence in what I can do and were always willing to improve what I was working on. Do it again? its plan to do a new internship during the summer 2015 with the Dessimoz lab for my master thesis, I am looking forward to doing this internship because I know I will work on a project that will provide me with new skills (biology, computer science or math) and that I’ll be mentored to produce something better.
•
Author: Ferenc Galkó •
∞
Note: this is the second interview in our new series “Life in the Lab”, which gives unedited accounts of students who have spent time with us. —Christophe
Please introduce yourself in a few sentences.
My name is Ferenc Galkó and I am a student from Hungary. I am 22 years old and will be graduating this December (2014) with a BSc in computer science from the Budapest University of Technology and Economics. I am really interested in doing an MSc and later possibly PhD abroad.
Why did you choose to join the lab?
As a BSc student I did not have much opportunities to contribute to the intriguing work of research labs, nor had I the time to move away from my home country for a longer period. I thought that an internship abroad would help to fulfil both of my aspirations.
What project did you work on (and for how long)?
I have worked for about three months on the migration of various operations on sequences, including a vectorized version of the Smith-Waterman algorithm written in C. The existing, efficient code base had to be migrated by using NumPy and Cython to make the main functionalities available from Python.
What came out of it?
At the end of the internship we had a new Python package called Python Optimal Pairwise Alignment (PyOPA), which provides an opportunity to perform operations with the efficiency of a vectorized C code with the convenience of a Python code. It is also a plan to publish the package in the PyPi shop in the very near future.
Was there any highlight or low point you’d like to share?
I had started my internship just a day before the lab retreat in Switzerland. I think it was a great thing to start the internship like this, because I have met almost everyone in person from the lab. During the retreat there was a special session addressed for pre-PhD students, where we discussed the advantages and the costs of doing a PhD. It was really great to hear unbiased advice from those who already went through this process or currently doing it.
What is your overall impression and would you do it again?
I would definitely do it again, it was a great and unique experience altogether. I have met interesting people from many countries and gained insight into a well-known research lab, which will surely form my upcoming years of study in a positive way.
•
Author: Leslie Macmillan •
∞
Note: this is the third interview in our new series “Life in the Lab”, which gives unedited accounts of students who have spent time with us. —Christophe
I just finished the M.Sc. Computer Science program at UCL, and did my summer project in the Dessimoz lab. I chose this lab because the project sounded interesting and the environment seemed supportive. Appearances turned out to be correct!
My project was increasing the speed of the existing Trees of Life program, which draws phylogenetic trees based on genetic distance within orthologous gene groups. At the end of the project, the overall speedup was 1.40X, and along the way I learned a lot about optimisation and many other topics: from UNIX-based systems to linear algebra to genetics.
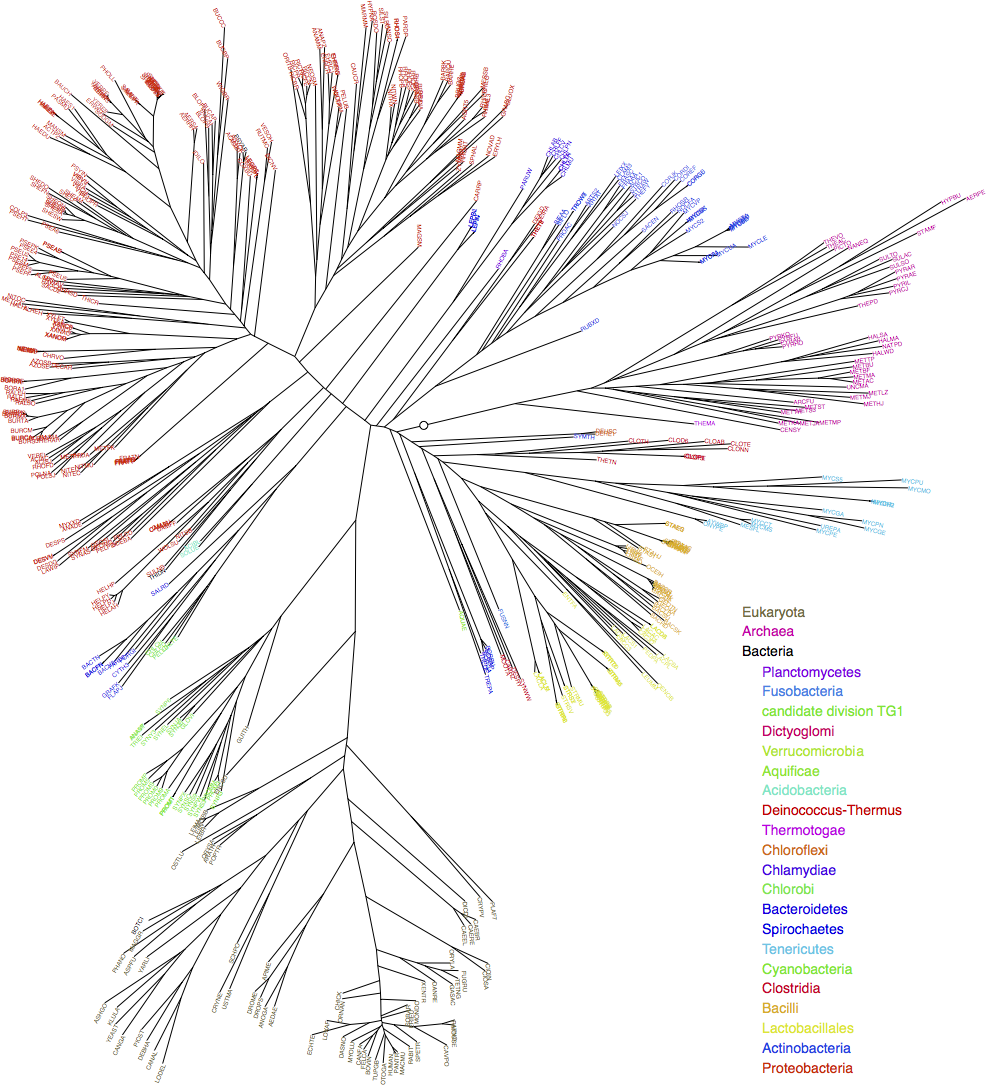
Sample output of the “TreeCollection” program
Overall I would really recommend working in this lab to anyone considering it. Though I was just here for three months, I felt very included and supported throughout the project. It was a great way to see the inner workings of a bioinformatics lab, and through this experience I was able to go to two conferences and a lab retreat in Switzerland!
 The Dessimoz Lab
blog is licensed under a Creative
Commons
Attribution 4.0 International License.
The Dessimoz Lab
blog is licensed under a Creative
Commons
Attribution 4.0 International License.