•
Author: Sina Majidian •
∞
Genomic data is expanding at a rapid pace, driven by ambitious efforts to sequence the DNA of millions of species worldwide. Comparative genomics, essentially the science of comparing genomes across species, helps us understand the evolutionary relationships between species. A key part of this is to find homologous regions, which are regions of DNA that are shared across species due to having a common ancestor.
When it comes to homologous genes, there are two main types to know about: orthologs and paralogs. Orthologs are genes that started diverging because of speciation (evolutionary branching into new species), while paralogs diverged because of gene duplication. Orthologs often have similar functions across species, which makes them extremely useful for transferring knowledge from well-studied organisms to newly sequenced ones (Nicheperovich 2022).
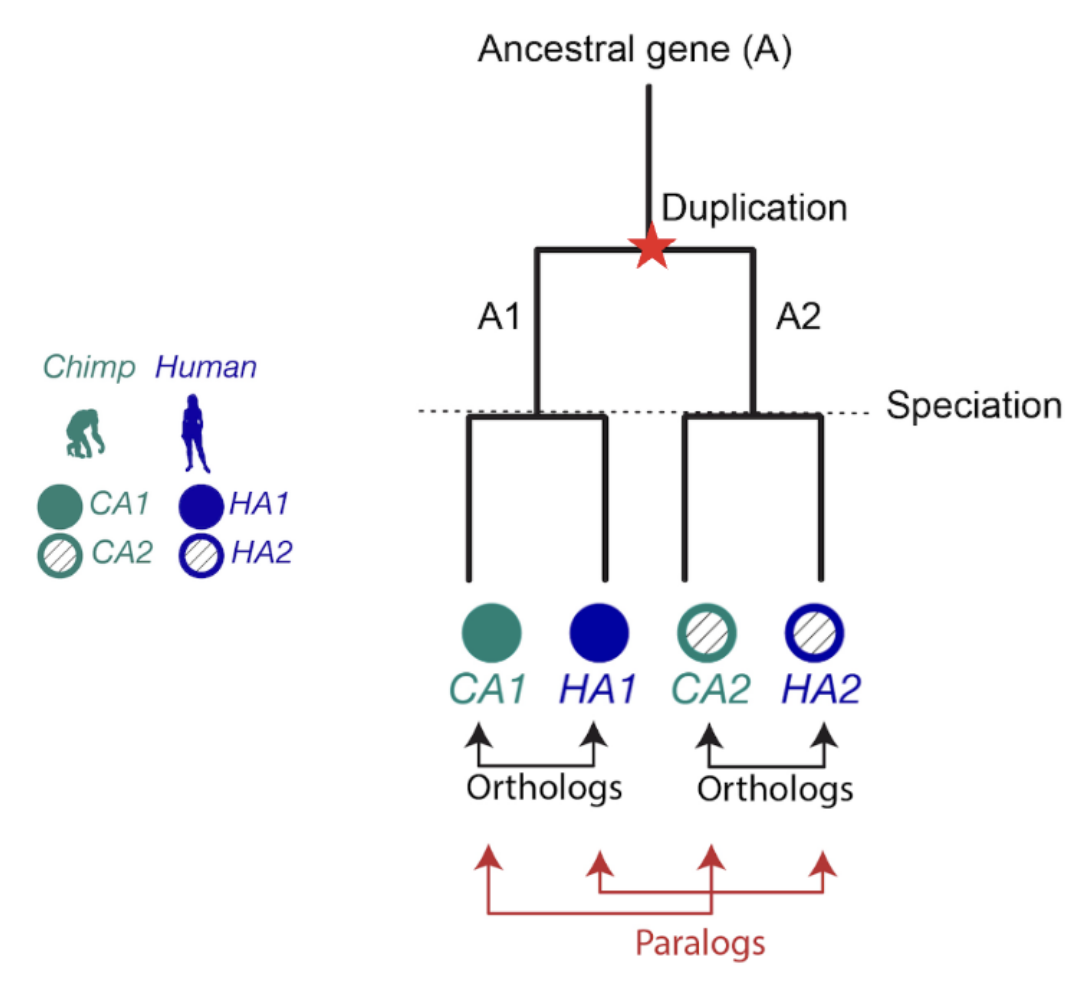
Figure 1. The relationship between two genes that share a common ancestor is called homologous, from the Greek word homologos— homos (meaning “same”) + logos (meaning “relation”). Orthologs are gene pairs that diverged due to evolutionary speciation, while paralogs are gene pairs that diverged due to a duplication event. This distinction is important because orthologs tend to have similar functions, but paralogs do not.
A bit of History!
The idea of distinguishing orthologs from paralogs goes back to Walter Fitch’s seminal work at the University of Wisconsin in 1970 (Fitch 1970). Since then, several research groups have been working on algorithms to accurately estimate orthology. One of the first contributions was the Clusters of Orthologous Groups of proteins (COGs) database, launched by NCBI in 2000, covering 21 genomes of bacteria, archaea, and eukaryotes (Tatusov 2000). More recently, the Orthofinder tool made it possible to find orthologs for a set of genomes of interest with high accuracy. This well-known software uses fast all-against-all gene comparisons with DIAMOND to group genes into orthogroups and refine them with gene trees. Earlier this year, Sonicparanoid presented its second version, which benefits from machine learning to efficiently avoid unnecessary all-against-all alignments, which makes it even faster. All these exciting advancements highlight the thriving community that works in the field of orthology and comparative genomics.
The OMA (Orthologous MAtrix) project came along in 2004 as a method and database for identifying orthologs across genomes (Dessimoz et al. 2005). The original OMA algorithm uses all-against-all gene comparisons with Smith-Waterman to find homologous sequences and then infers orthology relationships from there. Since 2010, Adrian Altenhoff has been the OMA project manager and OMA is hosted at the Comparative Genomics lab, led by Christophe Dessimoz and Natasha Glover. In 2017, Clément Train, a talented PhD student in the lab, took things to the next level with OMA algorithm 2.0, which delivered high precision in orthology inference (Train et al. 2017). Fast forward to today, the OMA Browser has seen 24 major updates where all the orthology data of around 3000 genomes is now presented for easy access with visualization innovations for phylostratigraphy, synteny and gene information (Altenhoff et al. 2024). Along the way, OMA also became a core resource supported by the SIB Swiss Institute of Bioinformatics.
In 2021, I joined the Comparative Genomics lab in Lausanne as a postdoc, took a leap of faith and started working on developing a new algorithm for orthology. The goal was to make it work for several thousands of species, basically scaling to the tree of life—something that’s really needed these days. At first, it felt quite overwhelming as there were several efficient ortholog inference tools such as Panther, OrthoMCL, Orthofinder, Sonicparanoid, Ensembl compara, Domainoid, MetaPhOrs, TOGA and GETHOGS (to name only a few) that are being maintained rigorously and regularly. The developer of these tools made great contributions to the field, and the huge number of comparative genomics studies over the years wouldn’t have been possible without these softwares. Their intricate design and comprehensive algorithms are accurate and efficient, making it hard to imagine advancing the field even further.
On top of that, I was new to the field—my PhD was on diploid and polyploid haplotype phasing using DNA sequencing reads (Majidian et al. 2020) and my background is in engineering and signal processing. But, I embarked on this journey and started learning concepts and methods in comparative genomics. I was lucky to have great mentors and lab mates who were always open to answering my questions, over zoom and in-person.
OMA turns young!
Let’s talk about FastOMA. With contributions from several lab members (Stefano, Yannis, Ali, Alex, David) and guidance from Christophe, Adrian and Natasha, we developed and implemented the FastOMA method. FastOMA works by benefiting from the current knowledge of orthology available on the OMA browser. FastOMA first maps the input genes (at amino-acid level) to reference gene families (the Hierarchical Orthologous Groups, HOGs), using OMAmer, a fast k-mer-based mapper. To learn about HOG, see this YouTube video by Natasha. Next, FastOMA works on each family separately. In other words, FastOMA does not perform comparison of genes from one family to another since these genes do not have any shared homology. This is an important step which saves us a huge amount of computations. Then, FastOMA infers the gene trees on (a subsample of) genes at each taxonomic level to distinguish orthologs from paralogs within each family. This phylogeny-guided subsampling is also key to maintaining speed and accuracy at the same time.
FastOMA’s speed makes it possible to handle genomic datasets with thousands of species. FastOMA uses the “OMA’s knowledge”, and is now swift as OMA turns young. FastOMA achieves high accuracy and resolution, as shown by the Quest for Orthologs benchmarks (Majidian, 2024).
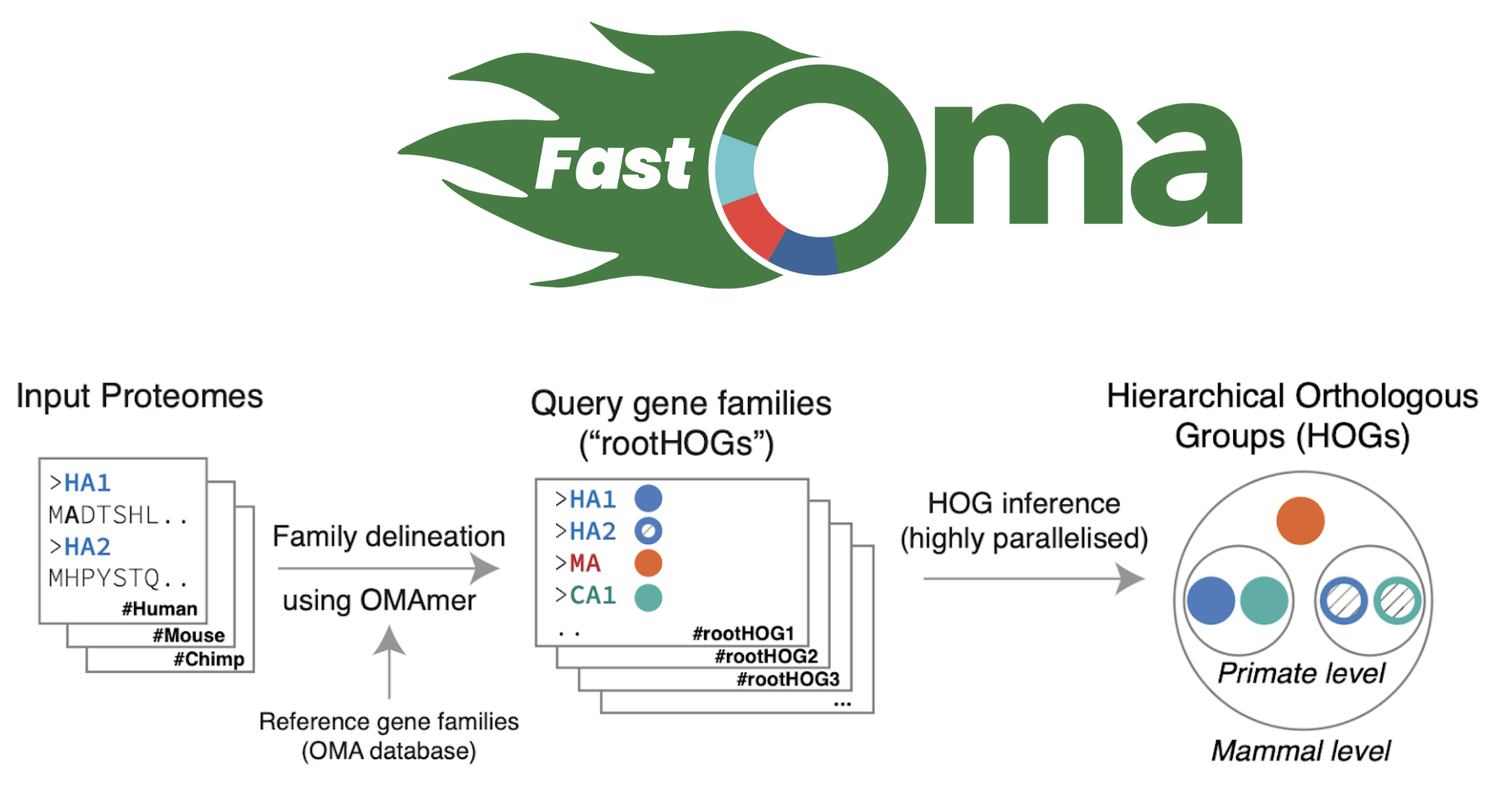
Figure 2. Overview of how FastOMA infers orthologs.
To the future!
As a community, we work collaboratively to advance the field and the lab has been contributing to the benchmarking datasets, making it possible to compare the performance of different tools, and ultimately advance the field. Earlier this year, in July, the Quest for Orthologs event (QFO8) was held at the University of Montreal, where recent advancements in orthology inference were discussed, and FastOMA was also presented there. The QFO 9 will be in Switzerland in 2026!
There are several directions for improving FastOMA’s accuracy and speed further. One exciting direction is taking advantage of recent advancements in protein structure prediction to reconstruct structural trees (Moi et al. 2023) in the context of orthology inference. This could really help boost resolution at deeper evolutionary levels. Besides, it would be very interesting to use gene order conservation, a.k.a, synteny information (Bernard et al. 2024), which could serve as an additional layer of information to refine orthology predictions. We hope our proposed hierarchical approach accompanied with several ideas will stimulate further developments.
So far, FastOMA has caught the attention of several labs around the world, who incorporated FastOMA in their studies. We are excited to hear how you plan to use FastOMA into your own research. Feel free to create a GitHub issue (https://github.com/DessimozLab/FastOMA) or send us an email if any help is needed!
To learn more see FastOMA academy: https://omabrowser.org/oma/academy/module/fastOMA
References
Altenhoff, Adrian M., et al. “OMA orthology in 2024: improved prokaryote coverage, ancestral and extant GO enrichment, a revamped synteny viewer and more in the OMA Ecosystem.” Nucleic Acids Research 52.D1 (2024): D513-D521. doi:10.1093/nar/gkad1020
Bernard, Charles, et al. “EdgeHOG: fine-grained ancestral gene order inference at tree-of-life scale.” bioRxiv (2024): 2024-08. https://doi.org/10.1101/2024.08.28.610045
Dessimoz, Christophe, et al. “OMA, a Comprehensive, Automated Project for the Identification of Orthologs from Complete Genome Data: Introduction and First Achievements” RECOMB 2005 Workshop on Comparative Genomics, LNCS 3678 (pp. 61-72). link
Emms, David M., and Steven Kelly. “OrthoFinder: phylogenetic orthology inference for comparative genomics.” Genome Biology 20 (2019): 1-14. doi:10.1186/s13059-019-1832-y
Fitch, Walter M. “Distinguishing homologous from analogous proteins.” Systematic zoology 19.2 (1970): 99-113. doi:10.2307/2412448
Majidian, Sina, et al. “Orthology inference at scale with FastOMA.” Nature Methods (2025) doi:10.1038/s41592-024-02552-8
Majidian, Sina, Mohammad Hossein Kahaei, and Dick de Ridder. “Minimum error correction-based haplotype assembly: Considerations for long read data.” PLOS ONE 15.6 (2020): e0234470. doi.org/10.1371/journal.pone.0234470
Moi, David, et al. “Structural phylogenetics unravels the evolutionary diversification of communication systems in gram-positive bacteria and their viruses.” BioRxiv (2023): 2023-09. doi:10.1101/2023.09.19.558401
Nicheperovich, Alina, et al. “OMAMO: orthology-based alternative model organism selection.” Bioinformatics 38.10 (2022): 2965-2966. doi:10.1093/bioinformatics/btac163
Tatusov, Roman L., et al. “The COG database: a tool for genome-scale analysis of protein functions and evolution.” Nucleic acids research 28.1 (2000): 33-36. doi:10.1093/nar/28.1.33
Train, Clément-Marie, et al. “Orthologous Matrix (OMA) algorithm 2.0: more robust to asymmetric evolutionary rates and more scalable hierarchical orthologous group inference.” Bioinformatics 33.14 (2017): i75-i82. doi:10.1093/bioinformatics/btx229
•
Author: Charles Bernard •
∞
For an evolutionary biologist, tracing today’s genomes back to key ancestors on the Tree of Life is a dream come true. With a collection of ancestral genomes, we could unravel the genetic steps that led to Life’s diversification from LUCA, the Last Universal Common Ancestor.
In practice, this means comparing modern genomes to find similar features—“orthologous” genes—passed down from common ancestors. By reversing this thinking, we can use these orthologous features as clues to “reconstruct” what ancestral genomes might have looked like.
But while much previous work has focused on reconstructing ancestral gene repertoires, reconstructing ancestral gene orders has been much more elusive.
In this post, I’ll dive into how we’ve developed a tool, EdgeHOG (1), to achieve this at a scale and speed never seen before (preprint here: https://www.biorxiv.org/content/10.1101/2024.08.28.610045v1).
Why do ancestral gene orders matter?
A genome isn’t just a random collection of genes; it has a structure that’s been shaped by evolution. Where a gene sits on a chromosome and its neighbours can matter a lot. Indeed, neighboring genes often work together (2). Plus, changes in gene order—genomic rearrangements—can lead to new traits and adaptations (3).
So, to understand the evolutionary history of these gene neighborhoods, we need to focus on gene adjacencies, not just the genes themselves. However, figuring out the gene order for every internal node on the Tree of Life is a huge computational challenge (4)…
How did we get into ancestral gene order inference?
When we wanted to analyze the link between gene function and gene adjacency across Life, we needed software that could reconstruct ancestral gene orders across the entire Tree of Life in one go, while accurately distinguishing between different copies of a gene in an ancestor.
But no tool could scale up to this level. Even the best tools, like AGORA (5) require reconstructing gene trees and perform pairwise comparisons of gene orders, which make them too slow to run on large datasets.
This is what drove us to create an algorithm with a clear goal: a linear time approach to reconstruct ancestral gene order, but without sacrificing accuracy.
How does EdgeHOG achieve linear-time complexity?
To infer ancestral gene orders at scale, our approach uses Hierarchical Orthologous Groups (HOGs). These model the lineage of genes from their ancestors to today’s species, assuming vertical inheritance.
By leveraging these gene lineages, our method uses “tree traversal” tricks to propagate or remove gene adjacencies along the species tree without any pairwise comparisons. Thanks to these tricks, our approach scales linearly with the size of the input phylogeny.
Since the software draws edges (gene adjacencies) between HOGs (proxies for ancestral genes), we called it EdgeHOG.
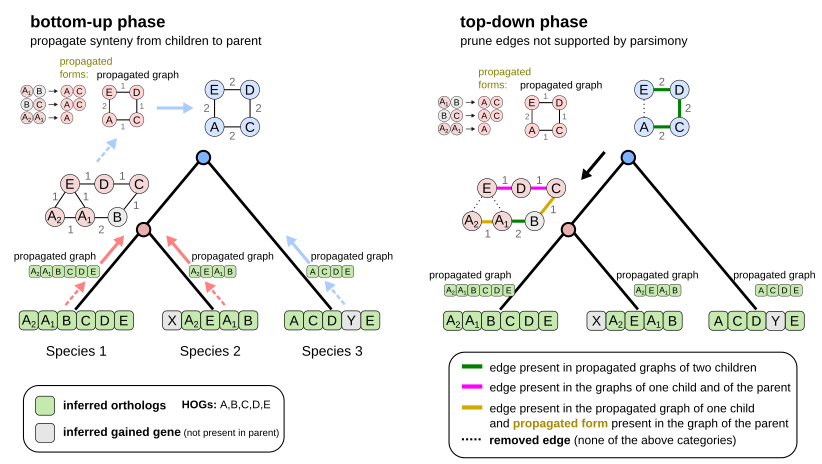
Here are the 2 first steps of EdgeHOG and the famous tree traversal tricks! The bottom-up phase propagates gene adjacencies up to the parental level of the species tree as long it is inferred by the HOG framework to have the two ancestral genes. The top-down phase essentially applies the Fitch algoritm and removes edges not supported by parsimony. Designing these tricks to comply with the constraint of linear time complexity was probably the most fun part of the project!
Fast and accurate
But EdgeHOG is not only fast, it is also very accurate! We validated it extensively on both simulated and real data. Across all benchmarks, EdgeHOG’s precision and recall met or exceeded the state of the art.
How to access EdgeHOG’s large scale inference of ancestral genomes?
The next step for us was to apply EdgeHOG to the entire OMA database, which currently includes 2,845 genomes from across the Tree of Life! This represent the first tree-of-life scale inference, resulting in 1133 ancestral genomes. You can explore these genomes on the OMA browser by clicking on Explore → Quick access to → Extant and ancestral genomes. For instance, check out the ancestral gene order for the last common ancestor of the mammals.
In the EdgeHOG paper, we also analysed the functions of the first ancestral contigs of genes ever reconstructed for LECA, the Last Eukaryotic Common Ancestor! These contigs contain genes that highlight core pathways like glycolysis, the pentose-phosphate shunt, amino-acid recycling, and histone organisation.
What kind of evolutionary analyses does EdgeHOG unlock?
In the lab, we’re using EdgeHOG to study the association between between gene order conservation and function conservation across different branches of the Tree of Life. We’re also dating gene adjacencies to identify old genomic neighbourhoods (like histone clusters in eukaryotes) or newer ones (like gene adjacencies on the sex chromosomes of animals).
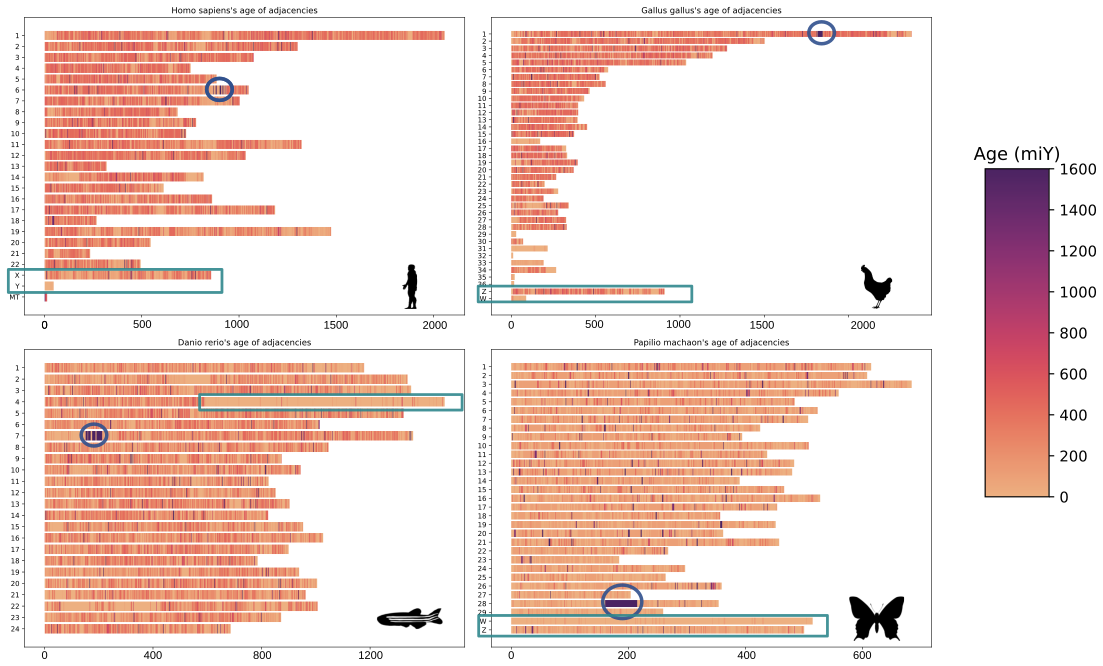
On these karyotypes, old histone clusters are circled in blue, and sex chromosomes are highlighted by rectangles. The estimated age of adjacencies is shown by the color scale on the right.
Overall, EdgeHOG opens up new possibilities in comparative genomics. For example, it helps track genomic rearrangements across a species tree, identify conserved gene clusters in clades of interest, or improve genome assembly by integrating gene order data from other species. Ultimately, knowing ancestral gene orders will enhance orthology inference by spotting highly divergent orthologs through their neighboring genes.
Do you want to try EdgeHOG on your datasets?
EdgeHOG is easy to use and available on GitHub https://github.com/DessimozLab/edgehog.
You’ll need a species tree, proteomes of each extant species (in Fasta files), and gene coordinates on contigs (in GFF files). Then, run our superfast FastOMA method to infer the HOGs. Finally, call EdgeHOG with the HOGs (OrthoXML file), the species tree (Newick file), and the path to the GFF files.
Now, you’re ready to perform big data ancestral gene order inferences, even with massive phylogenies of over 1,000 species! Try it on your favorite clade and let us know how it goes!
References
Bernard C, Nevers Y, Karampudi NBR, Gilbert KJ, Train C, Warwick Vesztrocy A, Glover N, Altenhoff A, Dessimoz C. EdgeHOG: fine-grained ancestral gene order inference at tree-of-life scale. bioRxiv 2024. https://doi.org/10.1101/2024.08.28.610045
Overbeek R, Fonstein M, D’Souza M, Pusch GD, Maltsev N. The use of gene clusters to infer functional coupling. Proc Natl Acad Sci U S A. 1999. https://doi.org/10.1073/pnas.96.6.2896
An X, Mao L, Wang Y, Xu Q, Liu X, Zhang S, Qiao Z, Li B, Li F, Kuang Z, Wan N, Liang X, Duan Q, Feng Z, Yang X, Liu S, Nevo E, Liu J, Storz JF, Li K. Genomic structural variation is associated with hypoxia adaptation in high-altitude zokors. Nat Ecol Evol. 2024. https://doi.org/10.1038/s41559-023-02275-7
El-Mabrouk N. Predicting the Evolution of Syntenies—An Algorithmic Review. Algorithms. 2021. https://doi.org/10.3390/a14050152
Muffato M, Louis A, Nguyen NTT, Lucas J, Berthelot C, Roest Crollius H. Reconstruction of hundreds of reference ancestral genomes across the eukaryotic kingdom. Nat Ecol Evol. 2023. https://doi.org/10.1038/s41559-022-01956-z
•
Author: David Moi •
∞
Breakthroughs don’t come every day, but the consequences of AlphaFold largely solving the 3D structure prediction problem has reshaped biology in profound ways. The sudden availability of protein structures for billions of proteins opens up many new possibilities. Last week’s two papers on the sequencing universe provide a compelling glimpse of the possibilities (here and here).
As someone who has been interested in tracing back the evolutionary origins of selected proteins—such as the cell fusion-mediating proteins fsx1 in plants, viruses, and archaea, or odorant receptors in insects—I have attempted to reconstruct phylogenies from structure in the past.
But I have faced two major issues:
Until AlphaFold came along, there typically wasn’t sufficient high-quality structure predictions as “starting material” to perform structure-based phylogenetics.
Even when I could obtain reasonably high confidence structures, the trees inferred from them were often met with skepticism—how reliable are these trees?
So now that high quality structure predictions are widely available, we could finally ask: are structures any good as starting material to infer trees? Specifically, how accurate are the reconstructed trees compared to sequences?
Today, we are super excited to report that structural phylogenetics works! What’s more, we found an approach that doesn’t just outperform traditional sequence-based methods for distant relationships; it also excels in resolving phylogenetic trees for closely related proteins. This post gives the gist of what we found—the full study is released as a preprint (1).
What’s the big deal with structural phylogenetics?
Before presenting our results, let’s take a step back. Why is structural phylogenetics potentially a big deal? Traditional phylogenetics, the study of evolutionary relationships among species or genes, has long relied on comparing the sequences of DNA, RNA, or proteins. While this approach has been immensely valuable, it does have its limitations. The primary challenge lies in the fact that the sequences of these biomolecules can change rapidly over time due to mutations and other factors, making it difficult to trace back their evolutionary history accurately when the divergence is very high. By contrast, proteins have unique three-dimensional structures that are intricately linked to their functions; these structures tend to change more slowly over evolutionary timescales compared to the sequences of the amino acids that make up the proteins since they are closely tied to the function of the protein.
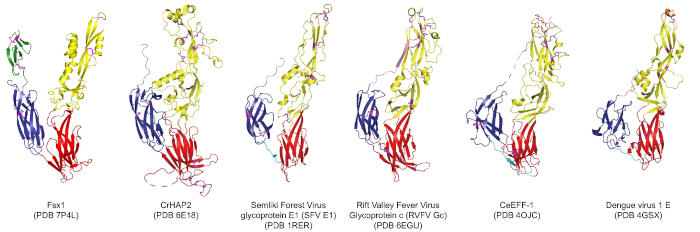
In this particular example close to my heart, we can see structural homology between functionally homologous proteins at wide evolutionary ranges. The examples shown span plants, metazoans, viruses and archaea. They share virtually no sequence homology. Ref: (2)
When we set out to do our work, however, we were not at all sure that it would work, let alone outperform sequence based methods. On the one hand, there have been decades of intensive tool and model refinements for sequence-based approaches, unlike its structure-based counterpart. But also, complications related to structure, such as allostery, flexible regions, and functional constraints could conceivably confound the evolutionary signal that can be extracted from structures.
Evidence that structure-based trees can outperform sequence-based trees
We tested a few structural approaches, and settled on an approach reconstructing distance trees using Foldseek’s “local structural alphabet” approach, which was developed in the lab of our collaborator Martin Steinegger to search for similar structures very rapidly—by encoding local structure motifs in a 20-letter alphabet and repurposing highly optimized alignment software originally developed to align amino acid sequences (3).
Testing and comparing the quality of phylogenetic trees empirically is tricky business. Most comparisons are based on simulated data, or by comparing the fit of data to different models. But how to compare trees that are reconstructed from entirely different kinds of input data? Luckily, our lab has accumulated quite some experience in these kinds of empirical observations, used previously to compare the accuracy of alignment (4 and 5) or orthology (6 and 7) methods. We used an approach which compares the propensity of inferred trees to recapitulate the known taxonomy of the species from which the proteins are sampled from.
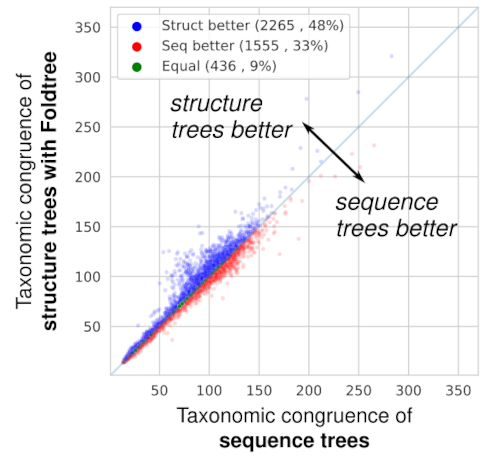
When comparing the taxonomic plausibility of thousands of trees derived from homologous protein families, Foldtree outperforms sequence-based phylogenetics. (In the paper, we show that after filtering the input set to families with high quality structures, the structural phylogenies perform even better!)
Amazingly, the trees we inferred in this way were more in line with the known taxonomy than those defined by sequence similarity! The input data can either be experimental crystal structures or AI structural models. Using good quality structures positively impacts the quality of the trees produced which means that as structural prediction methods get better, so will our structural trees.
The RRNPPA family: a first unifying phylogeny for peptidic quorum sensing proteins
To put our method to the test, we focused on a particularly complex gene family - the RRNPPA quorum sensing receptors (8). These receptors play a pivotal role in enabling communication and coordination among gram-positive bacteria, plasmids, and bacteriophages for crucial behaviors like sporulation, virulence, antibiotic resistance, conjugation, and phage lysis/lysogeny decisions.
The complex evolutionary pattern of this family is revealed in its name. Before AI structures, new homologs were previously only detectable after having been crystallized and each subfamily was added piecemeal to the overall picture, resulting in their particularly long acronym. As the family expanded researchers also attempted to piece together its evolutionary history, using a diverse set of methods, some of which relied on structural analysis. Using Foldtree we decoded the evolutionary diversification of these genes, shedding new light on their intricate history.
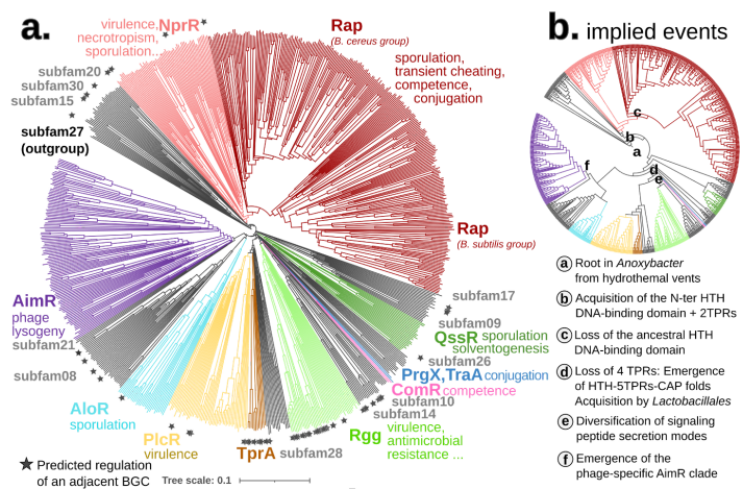
Compared to the sequence-based phylogeny, the Foldtree reconstruction of the RRNPPA family’s history is remarkably parsimonious. Several events such as domain architecture changes or transfers to the viral world appear only once in the tree.
Foldtree: infer a structural phylogeny for your favorite protein family

To make it easy to try this approach, as well as facilitate methodological improvements, we are releasing this new approach as an open source tool we call Foldtree. It’s available for download on GitHub (https://github.com/DessimozLab/fold_tree). Try it on your favorite protein family and let us know how it performs!
Exciting new research directions
High-accuracy structural phylogenetics has the potential to uncover deeper evolutionary relationships, elucidate unknown protein functions, and even refine the design of bioengineered molecules. The evolutionary histories of protein families in the viral domain, the start of eukaryotic life and the role of asgard archaea as well as the evolution of the prokaryotic mobilome are just a few cases where the fast pace of evolution has confounded sequence-based analyses and could be revisited. We believe this work represents an important step in investigating how structures are polished by the processes of evolution and how we can use this signal to peer further into the past than ever before.
References
Moi D, Bernard C, Steinegger M, Nevers Y, Langleib M, Dessimoz C. Structural phylogenetics unravels the evolutionary diversification of communication systems in gram-positive bacteria and their viruses. bioRxiv 2023.09.19.558401; doi: https://doi.org/10.1101/2023.09.19.558401
Moi D, Nishio S, Li X, Valansi C, Langleib M, Brukman NG, et al. Discovery of archaeal fusexins homologous to eukaryotic HAP2/GCS1 gamete fusion proteins. Nat Commun. 2022;13: 3880. doi:10.1038/s41467-022-31564-1
van Kempen M, Kim SS, Tumescheit C, Mirdita M, Lee J, Gilchrist CLM, et al. Fast and accurate protein structure search with Foldseek. Nat Biotechnol. 2023. doi:10.1038/s41587-023-01773-0
Tan G, Gil M, Löytynoja AP, Goldman N, Dessimoz C. Simple chained guide trees give poorer multiple sequence alignments than inferred trees in simulation and phylogenetic benchmarks. Proceedings of the National Academy of Sciences of the United States of America. 2015. pp. E99–100. doi:10.1073/pnas.1417526112
Dessimoz C, Gil M. Phylogenetic assessment of alignments reveals neglected tree signal in gaps. Genome Biol. 2010;11: R37. doi:10.1186/gb-2010-11-4-r37
Altenhoff AM, Dessimoz C. Phylogenetic and functional assessment of orthologs inference projects and methods. PLoS Comput Biol. 2009;5: e1000262. doi:10.1371/journal.pcbi.1000262
Altenhoff AM, Boeckmann B, Capella-Gutierrez S, Dalquen DA, DeLuca T, Forslund K, et al. Standardized benchmarking in the quest for orthologs. Nat Methods. 2016;13: 425–430. doi:10.1038/nmeth.3830
Bernard C, Li Y, Lopez P, Bapteste E. Large-scale identification of known and novel RRNPP quorum sensing systems by RRNPP_detector captures novel features of bacterial, plasmidic and viral co-evolution. Mol Biol Evol. 2023. doi:10.1093/molbev/msad062
•
Author: Yannis Nevers & Christophe Dessimoz •
∞
Proteins are fundamental to all life forms, dictating the complex biochemical interactions that maintain and drive the existence of every species. The functionality of a protein hinges on its structural domain organization, and the protein’s length is a direct manifestation of this. Given that every species has evolved under varying evolutionary pressures, one would intuitively expect protein length distribution to differ significantly across species.
Well, we report in a paper just published in Genome Biology that this is not the case.
Unexpected Homogeneity in Protein Length Distribution
In our study, we examined the protein length distribution across 2,326 species encompassing 1,688 bacteria, 153 archaea, and 485 eukaryotes. Counter to expectations, we observed a striking consistency in protein length distribution across these species. Though eukaryotic proteins were somewhat longer, the variation in protein length distribution was notably low compared to other genomic features such as genome size, gene length, number of proteins, GC content, and isoelectric points of proteins.
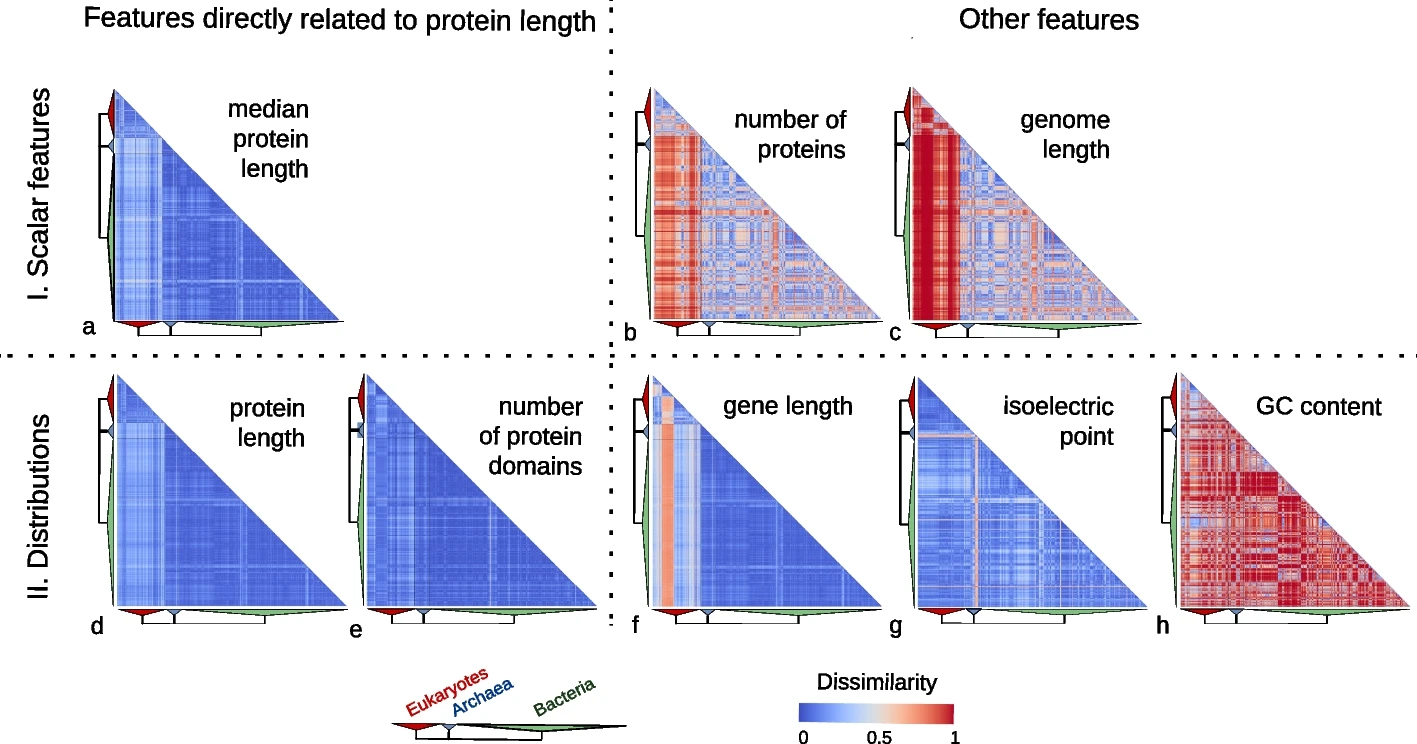
Features directly related to protein length are much more conserved than other features.
Exceptions: Errors or Biological Peculiarities?
We did note a few atypical cases of protein length distribution, but these were typically due to inaccuracies in gene annotation: no well-annotated model species displayed enrichment in small proteins, and those with a high number of small proteins were more likely to have incomplete or fragmented genome annotations.
Indeed, the outliers tended to include many more genomes scoring low in BUSCO quality score. The only exception we observed was the prevalence of longer proteins in the Ustilago fungal genus and the Apicomplexa phylum, known for their intracellular parasitic lifestyles.
This suggests that the actual variation in protein length distribution might be even smaller than what we reported. Hopefully, resequencing and reannotation efforts will help solve this issue in the future: we already noticed a few species getting updated proteomes where the length distributions gets more similar to the typical one!
A Universal Selection Force at Play
The startling uniformity of protein length distribution across diverse species suggests a strong, universal selective pressure, maintaining a high proportion of the coding sequence within a specific length range. In the discussion part of the paper, we articulate a number of potential explanations, but these remain highly speculative.
More positively put, the evolutionary forces behind the uniformity of protein distribution and their potential impact on fitness remain exciting areas of exploration!
Protein Length Distribution: A New Criterion for Gene Quality?
This observation led us to propose the use of protein length distribution as a new criterion of protein-coding gene quality upon publication. Considering that the overabundance of spurious proteins could potentially bias downstream analyses, this quality measure could aid in identifying and rectifying annotation errors. We also encourage everyone to take a look at this simple criterion when selecting proteomes for comparative genomics analysis.
Story behind the paper
The basic premise of the paper, exploring protein length distribution across the tree of life, may seem straightforward at first glance. Not quite. It started as part of Yannis’s PhD in Odile Lecompte’s lab in Strasbourg—and a few questions: what are the characteristics of the thousands of publicly available proteomes? How to decide which to include in large scale analyses? It took another three years of Yannis’s postdoc, with about half of that time spent in the peer-review process.
Perhaps the most revealing testament to the depth of this work is the supplementary PDF, a 68-page document filled with detailed data and analyses. Moreover, anyone interested in the peer-review history of our paper can delve into the 18-page record available here.
The journey is the reward, they say; well in this instance, we are quite happy to have reached our destination!
Reference:
Nevers, Y., Glover, N.M., Dessimoz, C, Lecompte, O. Protein length distribution is remarkably uniform across the tree of life. Genome Biol 24, 135 (2023). https://doi.org/10.1186/s13059-023-02973-2
•
Author: Christophe Dessimoz & Fritz Sedlazeck •
∞
We just published a method to build phylogenetic trees directly from raw reads, bypassing time-consuming steps such as genome assembly. This post gives the short story and the backstory. In particular, find out below what Read2Tree has in common with “Smoke on the Water” from the band Deep Purple.
In biology, phylogenetic trees are everywhere. They help us understand the relationships between species, genes, or cells—how they evolved, and how they’re related.
The sequencing revolution provides the raw material to infer phylogenetic trees, but building state-of-the-art phylogenetic trees requires tedious steps from read curation, de novo assembly, gene annotation, ortholog identification to tree inference, which can take many months to run—millions of CPU hours invested in this process are not uncommon—and specialised knowledge to oversee this process.
That’s where Read2Tree comes in. Our new approach to tree inference bypasses the usual steps of genome assembly, annotation, and orthology inference. Instead, it uses existing knowledge of the protein sequence universe to directly reconstruct comprehensive sequence alignments from raw sequencing reads.
The approach is vastly faster than traditional methods and in many cases more accurate—the exception being when sequencing coverage is high and reference species very distant. Read2Tree is also flexible, working with genome and transcriptome, short and long reads, and sequencing coverage as low as 0.1x.
We were encouraged by the buzz the Read2Tree manuscript elicited on bioRxiv last year, and are delighted it has now been published in Nature Biotechnology.
What is Read2Tree good for?
A nice illustration of Read2Tree’s potential was the reconstruction of a phylogeny of coronaviruses, which processed on the same tree diverse Coronaviridae sequences as well as 10,000 raw SARS-CoV-2 datasets from the Short Read Archive. The reconstructed tree was consistent with the lineage classification obtained from the UniProt reference proteomes, accurately recovering the main coronavirus genera and all subgenera (Figure 1). At the same time, the same phylogeny accurately clustered the sequences according to CDC variants of concerns classification. These results demonstrate the versatility and scalability of Read2Tree, making it suitable for both zoonotic surveillance and human epidemiology.
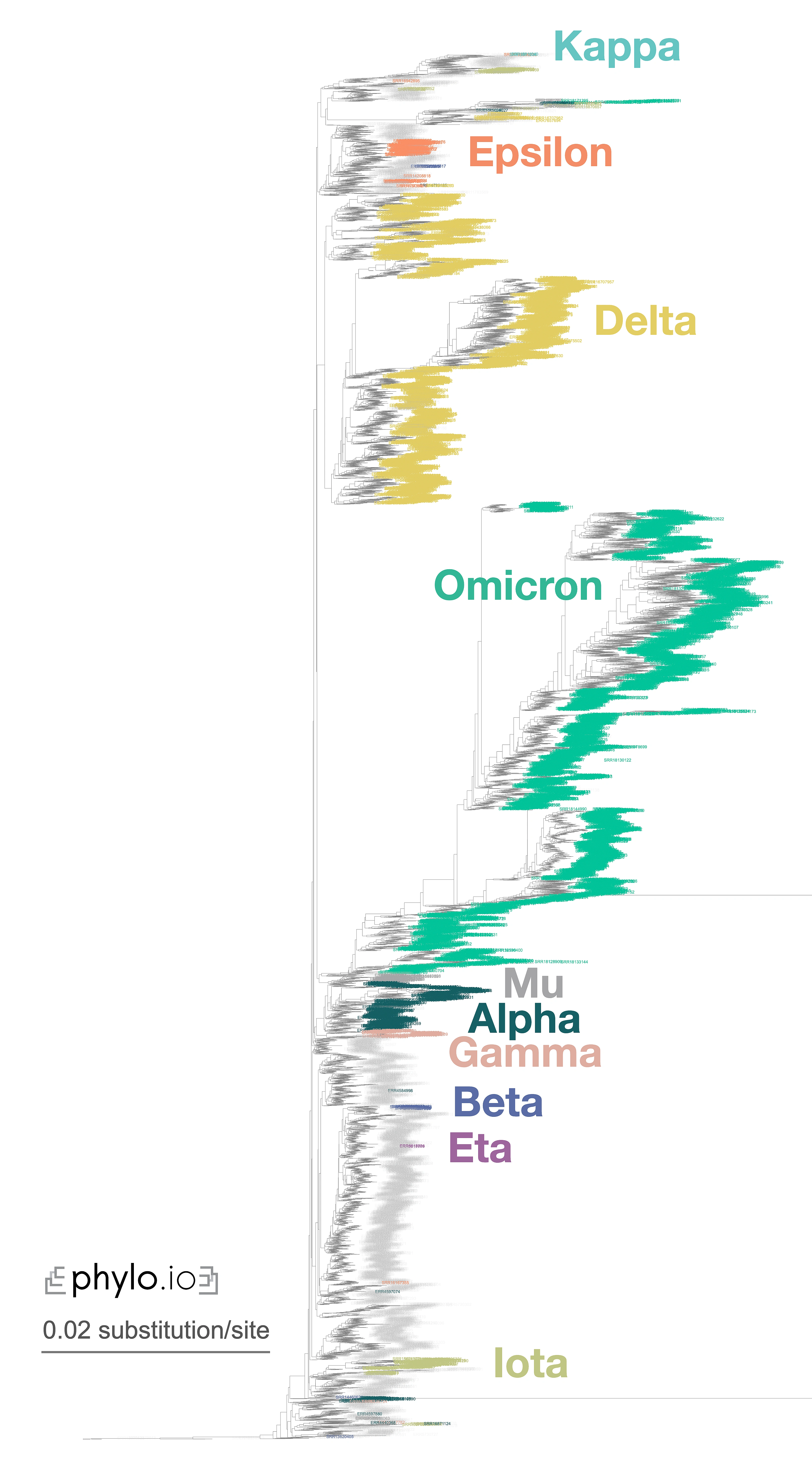
Figure 1—Zoomed-in display of a tree inferred using Read2Tree on 10,283 samples whole genome SARS-CoV-2 samples. Classification in colour was obtained from [https://harvestvariants.info](https://harvestvariants.info), where grey leaves are unclassified according to the CDC label. The colour clustering shows that the Read2Tree-based tree recovers consistent classification. Click on the tree to see it full screen.
The ability to reconstruct phylogenetic trees from raw reads has additional advantages. Some genomes are deposited with poor or even entirely absent protein annotation sets. Processing genomes directly from raw reads can avoid this limitation and also decrease biases that arise from relying too heavily on specific reference genomes. Although some efforts have been made to “dehumanize” non-human great ape genomes, other clades still face similar biases that can be significantly reduced by processing raw reads.
Who might find it useful?
We think Read2Tree will be especially useful for small labs with limited bioinformatics expertise and computational resources, allowing them to perform state-of-the-art phylogenomics on particular species or environments of interest.
But it’s not just small labs that can benefit from Read2Tree. Large consortia can also use it to regularly update their trees as new genomes are sequenced. This is especially important as more and more projects around comparative genomics are underway, such as the Earth BioGenome, the Darwin Tree of Life, or the European Reference Genome Atlas projects.
In addition, Read2Tree’s ability to infer trees from much lower coverage than traditional methods means it can also be useful for quality control early in the process. This makes it a valuable tool for environmental and metagenomic applications, especially when combined with genome binning techniques.
Overall, Read2Tree is a powerful method for inferring phylogenetic trees directly from raw sequencing reads. We hope it will help make phylogenetic tree inference faster, more accurate, and more accessible to a wider range of researchers.
What’s next?
Now that the introductory Read2Tree paper is published, we are excited to explore new potential applications that we haven’t been able to tackle so far. For instance, we have already received inquiries from researchers interested in using Read2Tree for ancient DNA applications or for monitoring systems that require fast turnaround time and low coverage.
Moving forward, we have two main goals. First, we aim to expand Read2Tree’s capabilities to handle multi-species samples, which will enable an even broader range of applications in the metagenomics field. While long-read applications may offer the most benefit, we are confident that Read2Tree’s ability to perform well with short-reads will also prove valuable in detangling multiple species.
Secondly, we plan to explore the use of Read2Tree in single-cell sequencing. This rapidly growing field involves sequencing individual cells, including cancer cells, and analysing their genetic information. Given Read2Tree’s ability to operate with low coverage levels (down to 0.2x), we believe it could facilitate fast and accurate characterization of tumour or cell evolution.
We hope that Read2Tree will help streamline and democratise comparative genomics analyses. We are excited to see how researchers will apply this tool to further advance our understanding of genetics and evolution.
What’s the backstory?
Both of our labs (Fritz Sedlazeck’s and Christophe Dessimoz’s) have been collaborating for many years, and we’ve always enjoyed exchanging ideas even though our research interests are quite diverse. One of our interests over the years is how to combine our expertise in sequence analysis and ortholog comparison to develop new methodologies and gain new insights into biology.
It was during one of Fritz’s visits to Christophe’s lab in Lausanne, Switzerland, that we started brainstorming ideas for a project that led to Read2Tree. Our goal was to overcome the limitations and bottlenecks of comparative genomics. We had some amazing cheese risotto, and the beautiful scenery fueled our discussions further (Figure 2).

Figure 2 — Fritz alleges that the epiphany of Read2Tree took place with this view from his hotel room in Montreux, Switzerland, during a collaborative visit to Christophe’s group. It’s not entirely implausible, considering this very view [inspired the song “Smoke on the Water” by Deep Purple](https://en.wikipedia.org/wiki/Smoke_on_the_Water#History).
David Dylus, the first author, was convinced that it was possible to bring our ideas to life, although he did not anticipate how much time and effort it would take (Figure 3). Even after he moved on to a new role in the pharmaceutical industry, he continued to work on Read2Tree after regular work hours. And when the COVID-19 pandemic hit, we had to face additional challenges, such as maintaining regular meetings and pushing the manuscript forward while not compromising on quality. We also faced technical issues, such as hard disk crashes and cluster updates that led to data loss, but David hang on.
Completing the paper was not an easy task, and one of the biggest challenges was organising and identifying all of the SRA data sets, including those related to yeast and COVID-19. Despite these challenges, we were able to bring the project to completion. It was a special joy to present the work at ISMB 2022, where Fritz and Christophe had the wonderful opportunity to meet in person, and we continued to discuss our work while enjoying good food and drinks by beautiful Mendota lake in Madison, Wisconsin.
In summary, nice food and lakeside views were instrumental in the making of Read2Tree.

Figure 3 — First author David Dylus performing on stage (centre, crouching) on the occasion of SIB Swiss Institute of Bioinformatics’s 20th anniversary—a period of rapid progress in the development of Read2Tree. Though no-one is entirely certain, rumour has it that David is miming “sipping a cup of tea while looking into the distance”, in line with our theme of sustenance, inspiring landscapes, and scientific progress.
Note this blog post was first published on the Nature Communities blog here.
Reference
Dylus, D., Altenhoff, A., Majidian, S. et al. Inference of phylogenetic trees directly from raw sequencing reads using Read2Tree. Nat Biotechnol (2023). doi:10.1038/s41587-023-01753-4.
•
Author: Nastassia Gobet •
∞
When I started using word processors, the spell checker was only looking at small and common typing errors and was often trying to correct acceptable words due to lack of vocabulary. A few years later, they not only are better at it and use more developed dictionaries, but they can also capture grammar mistakes and redundant phrases. A similar story is happening with the detection of genomic variants.
The genome as a big text
The genome can be considered as a big text, written in a 4-letter alphabet (A, C, G, T). When comparing the genomic words from two individuals, we can look at single or few letter(s) differences (single nucleotide variants, SNVs) and longer patterns (structural variants, SVs) such as words, sentences, and paragraphs that are added (insertions) or missing (deletions), exchanged (translocations), repeated (duplications and copy number variations, CNVs), inverted (inversions) or combinations of these (complex SVs).
Discovering the importance of SVs
About ten years ago, the focus was mainly on SNVs as these are numerous and many methods to detect them were developed. They were studied in deep and indexed in dictionaries (databases) that also document their frequencies. However, one letter differences do not necessarily have a significant effect on the meaning of the text (the phenotypes). On the other hand, although SVs were underestimated and consequently understudied, they were discovered to have a profound phenotypic impact on gene regulation, dosage, and function. Therefore, they are important in a wide variety of medical conditions: cancers, neurological diseases (Parkinson, Huntington), and mental disorders (autism, schizophrenia).
Challenges in SV identification
Methods were recently developed and are currently being developed to detect SVs. A number of challenges need to be dealt with. First, short read sequencing greatly limits the detection of large events exceeding read length. Consequently, using longer read technologies (PacBio and ONT) is improving the range of detectable SVs, but this comes at the cost of decreased sequencing accuracy and higher price. Hybrid strategies combining short and long reads are therefore promising. Second, SVs are hard to classify as the variant type depends on variant sequence context: a sequence can be considered an insertion, duplication, or translocation depending on the source (Figure 1). In addition, the number of possible SVs is infinite, whereas for SNVs there are 3 variants per position in the worst case. SVs are thus hard to compare: which criteria should we use to determine if two slightly different calls correspond to the same event or not? This affects SV reporting and frequencies. Due to the relative youth of the field, standards and best practices have yet to be established. Different initiatives (eg. Genome in a Bottle and SEQC2) aim at better characterizing false positives and false negatives in SV calling. This should help implement more objective benchmarking and comparison between the various detection methods.
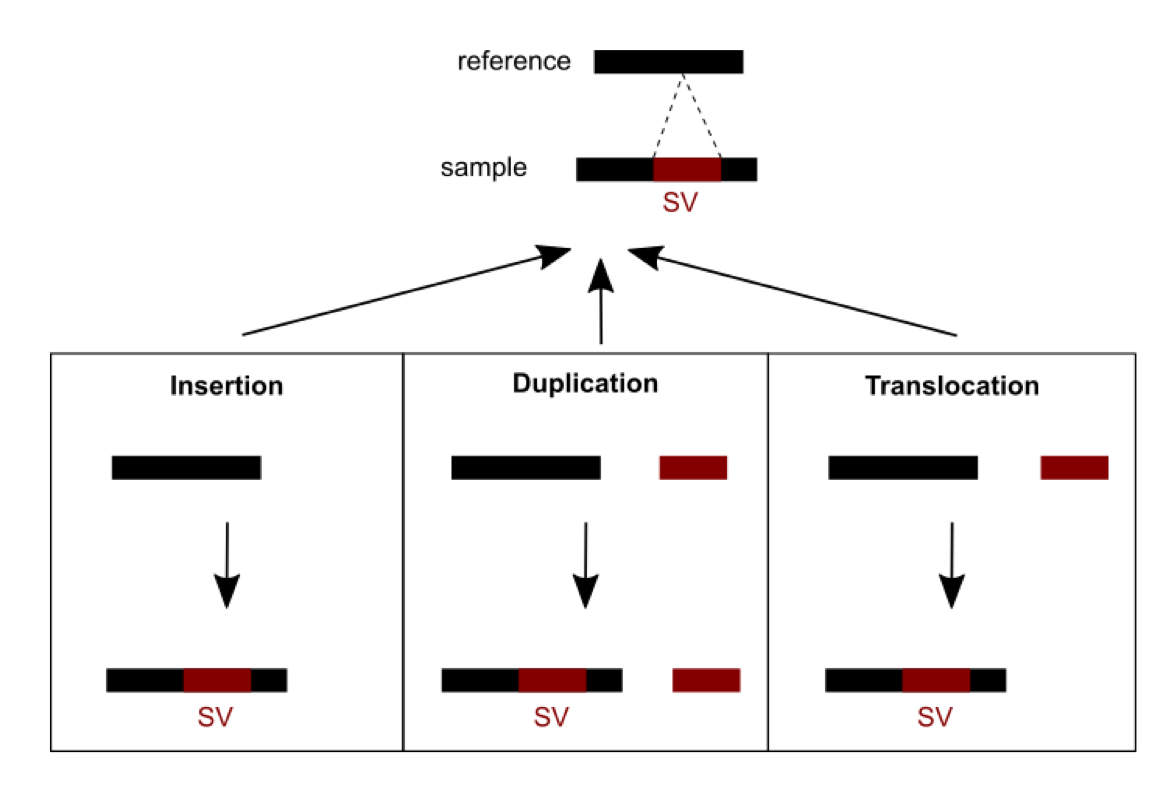
Figure 1: An SV was called for a sequence from a sample differing from the reference sequence. Three possible scenarios of formation could explain the SV observed: an insertion, a duplication or a translocation.
Future of genomic spelling and grammar checkers
Standards and objective benchmarking for SV detection are still missing, so one must be careful with results obtained from current methods. However, SVs are increasingly recognized as being important and technologies to detect them are evolving rapidly. I think their use will become a more common practice in genomic variation studies in a few years, similar to spelling and grammar checkers in text processors. And you, which genome checker will you use?
Reference
Mahmoud M, Gobet N, Cruz-Dávalos DI, Mounier N, Dessimoz C, Sedlazeck FJ. 2019. Structural variant calling: the long and the short of it. Genome Biol 20:246. doi:10.1186/s13059-019-1828-7.
If you want to get involved in improving SV variant detection, consider joining this Hackathon, to be hold remotely Oct. 11-14, 2020.
•
Author: Christophe Dessimoz •
∞
The problem in a nutshell
Quantitative Trait Loci (QTL) are regions of a genome for which genetic variants correlate with particular traits. To take a simple example in plants, one might observe that the average seed size (trait) is significantly larger when considering the subset of a population which has a C at a particular position in the genome than a subpopulation with a T.
The reason QTL identifies genomic regions and not precise positions is that neighbouring variants tend to be inherited together. These regions typically contain hundreds of genes, making it difficult to say which one(s) are causal to the trait variation—if any at all (the causal genetic variation(s) can be in non-coding regions too).
Thus, to prioritise candidate causal genes within a QTL region, researchers typically consider previous knowledge on these genes, to see whether a particular gene “makes sense”. In the case of seed size, it might be a gene previously implicated in growth or regulation, or a gene known to influence seed size in a different species. This process is however requires substantial manual interpretation, and is thus labour-intensive and haphazard.
Enter QTLsearch
We realised that our framework of hierarchical orthologous groups, which relates genes across many species, could be extended to integrate QTL results with previous gene function annotations.
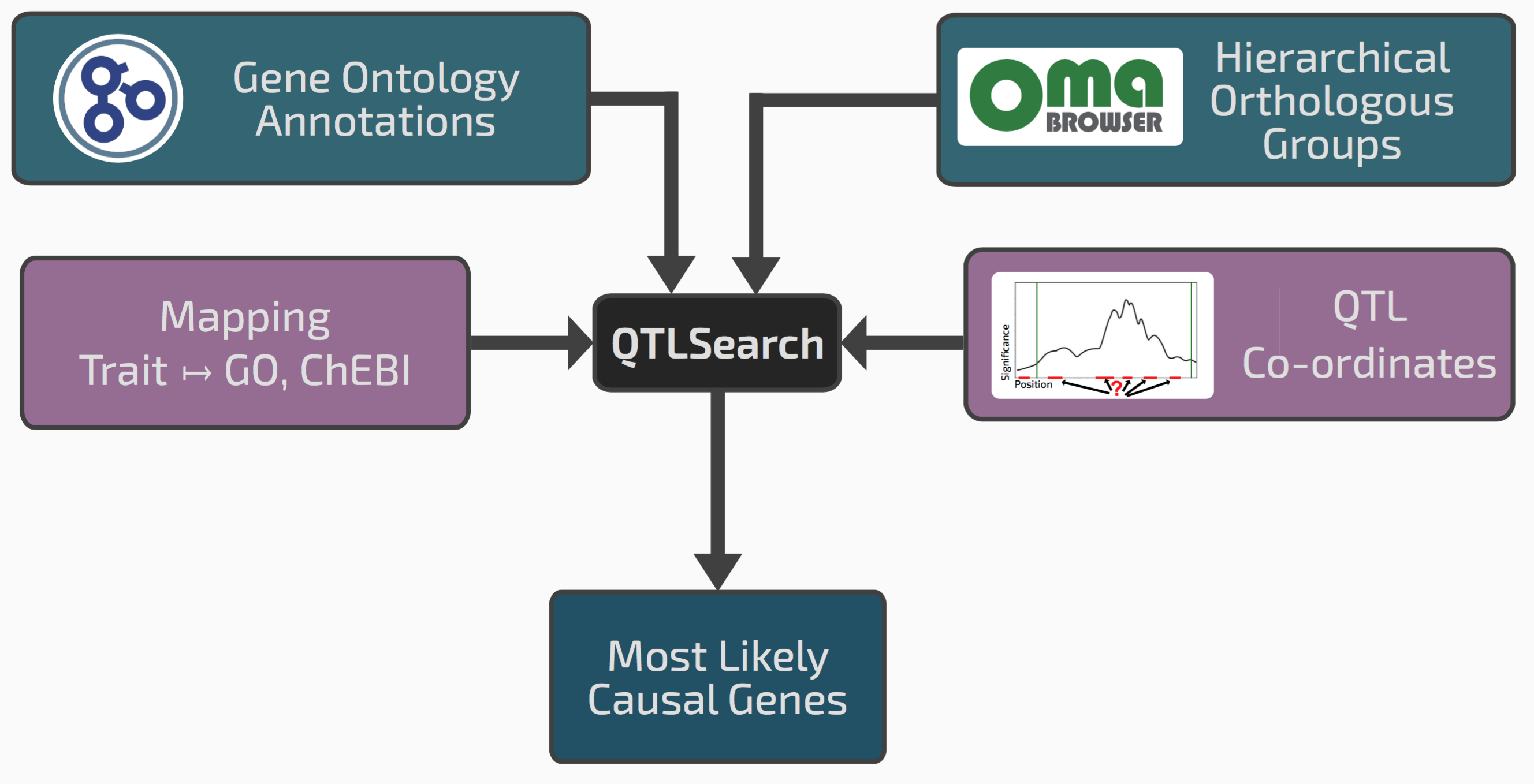
Conceptual overview of QTLsearch
If we go back to the seed size example, it might be that among the genes in the window, one has an ortholog in a different species previously annotated with the GO term “reproductive system development”. This could be a good candidate causal gene.
One risk however in integrating lots of previous knowledge across many species is that we might also find some spurious patterns. We therefore had to devise a way of controlling for random associations between QTL regions and evolutionarily propagated knowledge. Such “null distribution” depends on the specificity or the terms in question, the amount of annotations, the size of the QTL regions, and the species sampling. To cope with this complexity, we chose to implement a non-parametric permutation test.
We implemented the tool as an open source package called QTLsearch, available here.
QTLsearch infers more candidate causal genes than manual analyses
We used QTLsearch to reanalyse two previous studies. In both cases, we could call more candidate genes than the original studies. But more importantly, the evidence behind our calls is fully traceable and statistically supported.
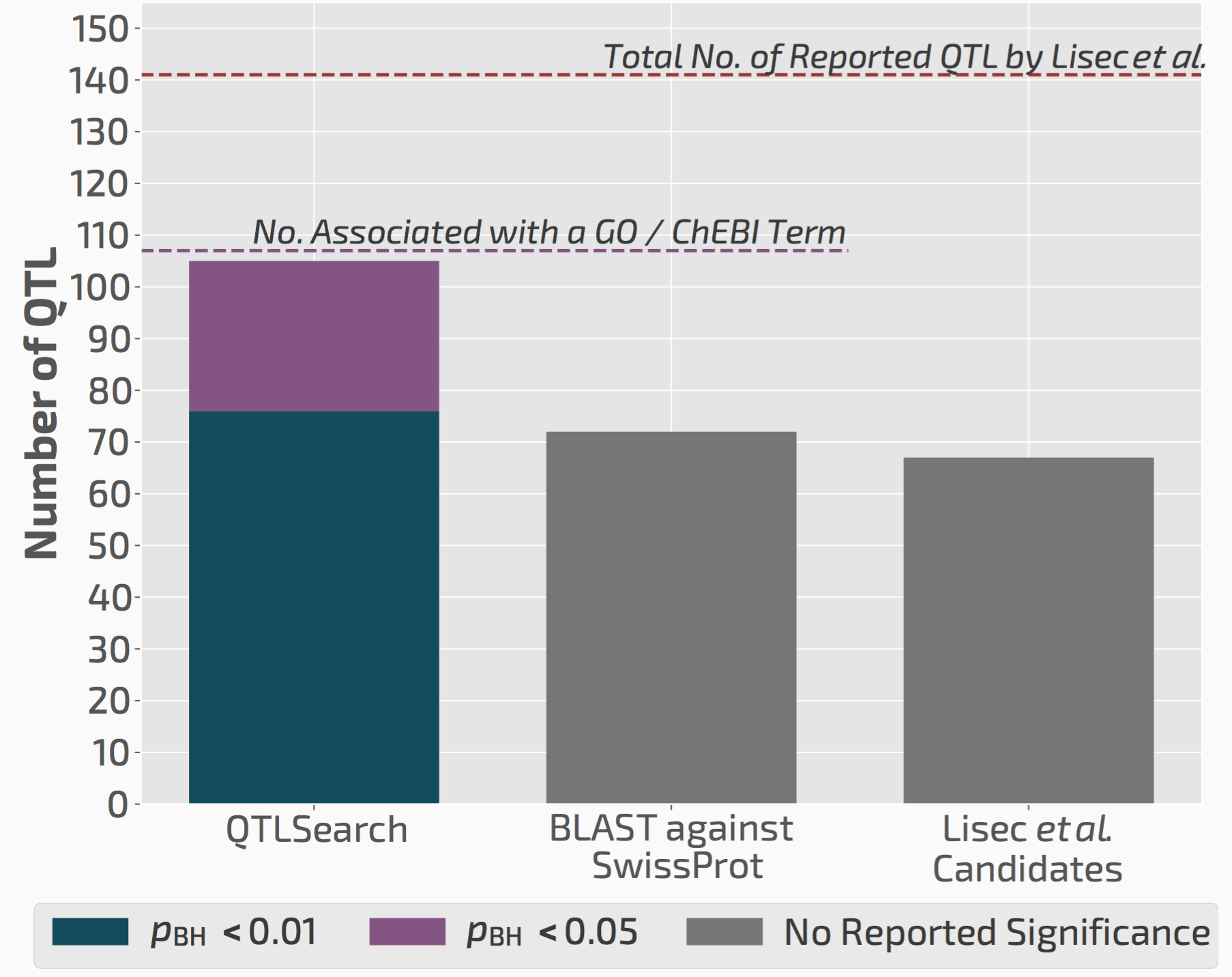
QTLsearch could identify more candidate genes than the original study, but in an automated, reproducible, and statistically meaningful way.
Thus we think this will greatly facilitate future QTL analyses, particularly those that are done in non-model species for which the previous experimental knowledge is very limited.
Behind the paper
This is the third paper that resulted from our collaboration with Bayer CropScience (now BASF CropScience), after our work on homoeologs and on detecting split genes.
The project was conceived by Henning Redestig, collaborator at Bayer at the start of the project (now at DuPont). Henning had contributed to a QTL study and knew how labor intensive the search for putative causal genes is. He realised that HOGs could provide a natural way of integrating functional knowledge across multiple species, to combine the QTL information with previous functional data.
Alex Warwick Vesztrocy, PhD student on the project and first author, ran with the idea—promptly implementing and testing it. Early results looked promising, but Alex soon realised that the mapping between metabolites and GO terms could be improved. He also realised that some terms were quite common, so he devised the approach to compute the significance scores.
Our manuscript was accepted as proceedings paper at the European Conference on Computational Biology (ECCB). In our lab, we like proceedings paper. It’s nice to be able to present the work and publish the paper, particularly since the ECCB proceedings appear in a good journal. More importantly, conferences impose hard deadlines. Deadlines for submission of course, but also for peer-reviewing and for deciding acceptance or not!
Reference
Alex Warwick Vesztrocy, Christophe Dessimoz*, Henning Redestig*,
Prioritising Candidate Genes Causing QTL using Hierarchical Orthologous Groups, Bioinformatics, 2018, 34:17, pp. i612–i619 (ECCB 2018 proceedings)
[Open Access Full Text]
•
Authors: Fritz Sedlazeck, Dan Jeffares & Christophe Dessimoz •
∞
Our latest study just came out (Jeffares et al., Nature Comm 2017). In it, we carefully catalogued high-confidence structural variants among all known strains of the fission yeast population, and assessed their impact on spore viability, winemaking and other traits. This post gives a summary and the story behind the paper.
Structural variants (SVs) measure genetic variation beyond single nucleotide changes …
Next generation sequencing is enabling the study of genomic diversity on unprecedented levels. While most of this research has focused on single base pair differences (single nucleotide polymorphisms, SNPs), larger genomic differences (called structural variations, SVs) can also have an impact on the evolution of an organism, on traits and on diseases. SVs are usually loosely defined as events that are at least 50 base pair long. They are often classified in five subtypes: deletions, duplications, new sequence insertions, inversions and translocations.
Over the recent years the impact of SVs has been characterized in many organisms. For example, SVs play a role in cancer, when duplications often lead to multiple copies of important oncogenes. Furthermore, SVs are known to play a role in other human disorders such as autism, obesity, etc.
… but calling structural variants remains challenging
In principle, identifying SVs seems trivial: just map paired-end reads to a reference genome, look for any abnormally spaced pairs or split reads (i.e. reads with parts mapping to different regions), and—boom—structural variants!
In practice, things are much harder. This is partly due to the frustrating tendency for SVs occur in or near repetitive regions where short read sequencing struggles to disambiguate the reads. Or in highly variable regions of genome such as the chromosome ends, which tend to be the tinkering workshop of the genome.
As a result, a large proportion of SVs—typically at least 30-40%—remain undetected. As for false discovery rates (proportion of wrongly inferred SVs), they are mostly not well known because validating SVs on real data is very laborious.
Fission yeast: a compelling model to study structural variants
Studying structural variants in Schizosaccharomyces pombe is especially suited because:
- The genome is small, well-annotated and simple (few repeats, haploid).
- We had 40x or more coverage over 161 genomes covering the worldwide known population of S. pombe.
- We had more than 220 accurate trait measurements for these strains at hand. Since the traits are measured under strictly controlled conditions, they contain little (if any) environmental variance—in stark contrast to human traits.
SURVIVOR makes the most out of (imperfect) SV callers
To infer accurate SVs calls, we introduced SURVIVOR, a consensus method to reduce the false discovery rate, while maintaining high sensitivity. Using simulated data, we observed that consensus calls obtained from two to three different SV callers could recover most SV while keeping the false-discovery rate in check. For example, SURIVOR performed second best with a 70% sensitivity (best was Delly: 75%), while the false discovery rate was significantly reduced to 1% (Delly: 13%) (but remember these figures are based on simulation; performance on real data is likely worse.) Furthermore, we equipped SURVIVOR with different methods to simulate data sets and evaluate callers; merge data from different samples; compute bad map ability regions (BED file) over the different regions, etc. SURVIVOR is written in C++ so it’s fast enough to run on large genomes as well. Since then, we are running it on multiple human data sets, which takes only a few minutes on a laptop. SURVIVOR is available on GitHub.
SVs: now you see me, now you don’t
We applied SURVIVOR to our 161 genomic data sets, and then manually vetted all our calls to obtain a trustworthy set of SVs. We then discovered something suspicious. Some groups of strains that were very closely related (essentially clonal, differing by <150 SNPs) had different numbers of duplications, or different numbers of copies in duplications (1x, 2x, even 6x). This observation was also validated with lab experiments.
Interestingly we identified 15 duplications that were shared between the more diverse non-clonal strains (so these must have been shared during evolution) but could not be explained by the tree inferred from SNPs (Figure 1). To confirm this we compared the local phylogeny of SNPs in 20kb windows up and downstream of the duplications with the variance in copy numbers. Oddly the copy number variance was not highly correlated with the SNP tree. This lead to the conclusion that some SVs are transient and thus are gained or lost faster than SNPs.
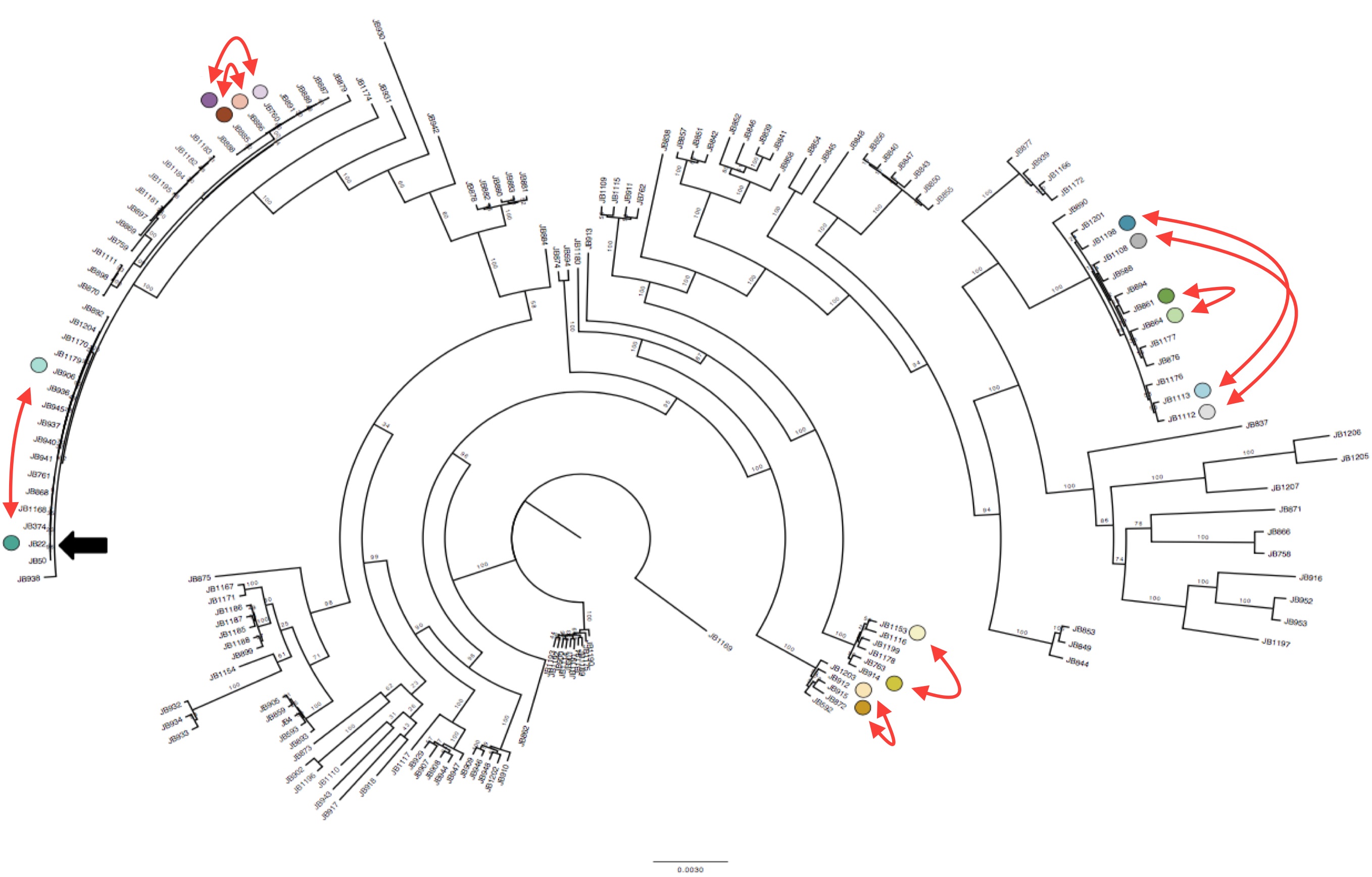
Duplications happen within near-clonal populations Phylogenetic tree of the strains reconstructed from SNPs data, with eight pairs of very close strains that nonetheless show structural variation. Click to enlarge.
Though this transience came as a surprise, there is actually supporting evidence from laboratory experiments carried out by Tony Carr back in 1989 that duplications can occur frequently in laboratory-reared S. pombe, and can revert. (Carr et al. 1989). The high turnover raises the possibility that SVs could be an important source for environmental adaptation.
SVs affect spore viability and are associated with several traits
We then investigated the phenotypic impact of these SVs. We used the 220 trait measurements from previous publications. We observed an inverse correlation between rearrangement distance and spore viability, confirming reports in other species that SVs can contribute to reproductive isolation. We also found a link between copy number variation and two traits relevant to wine making (malic acid accumulation and glucose+fructose ultilisation) (Benito et al. PLOS ONE 2016).
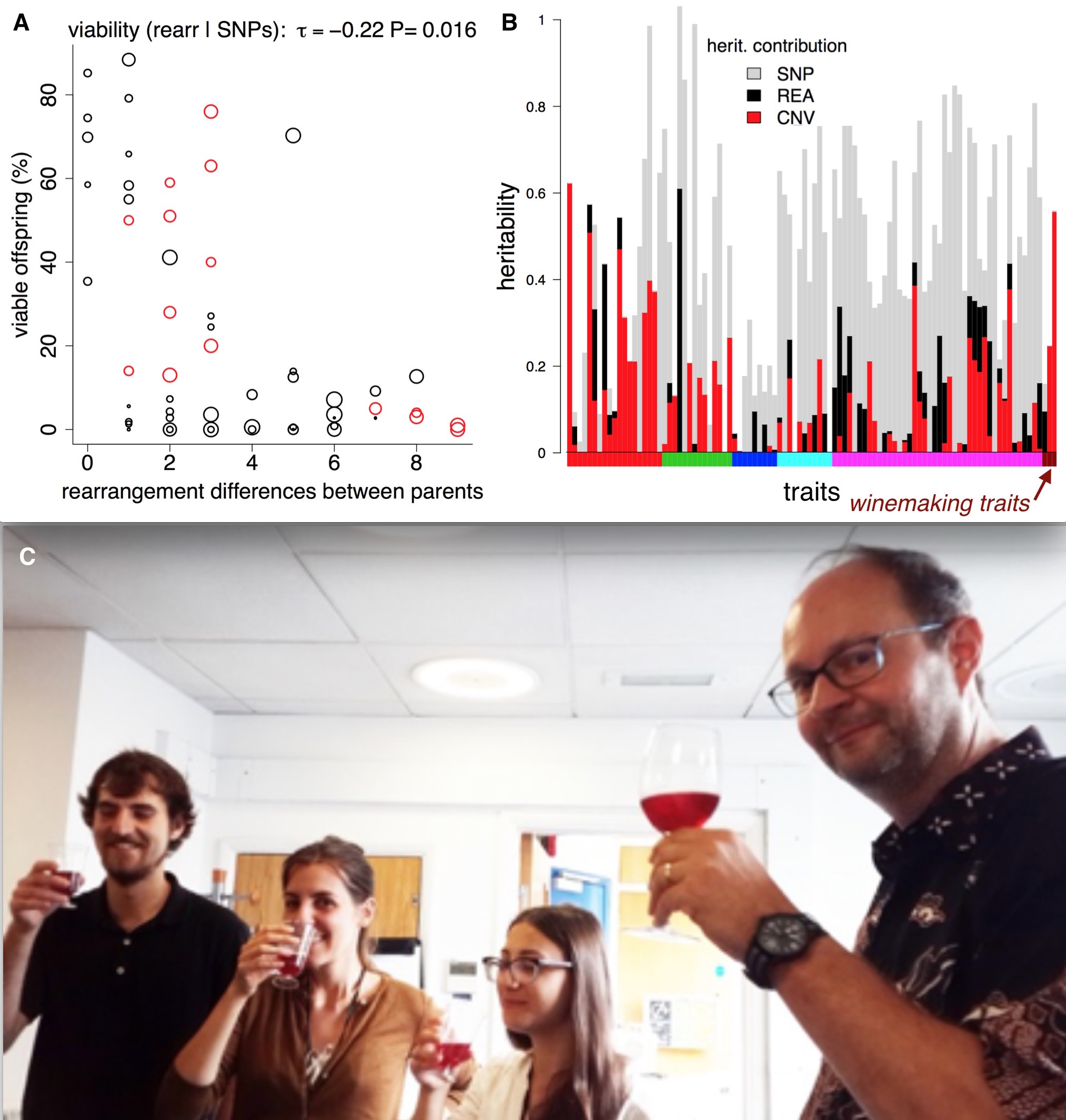
Structural variants, reproductive isolation, and wine. A) Making crosses between fission yeast strains often results in low offspring survival. The theory is that rearrangements (inversions and translocations) cause errors during meiosis, so we might expect them to affect offspring viability. If we compare offspring viability from crosses with the number of rearrangements that the parents differ by, there is a correlation, and a ‘forbidden triangle’ in the top right of the plot (it seem impossible to produce high viability spores when parents have many unshared rearrangements). B) SVs also affect traits. For > 200 traits (vertical bars) we used [LDAK](http://dougspeed.com/ldak/) to estimate the proportion of the narrow sense heritability that was caused by copy number variants (red), rearrangements (black) and SNPs (grey). Some traits are very strongly affected by copy number variants, such as the wine-making traits (wine-colored bars along the x-axis). C) Fission yeast wine tasting at UCL—how much of the taste is due structural variants? (Jürg Bähler at right).
We used the estimation of narrow sense heritability from Doug Speed’s LDAK program. Narrow sense heritability estimates how much of a difference in a trait between individuals can be explained by adding up all the tiny effects of the genomic differences (in our case SNPs; deletions and duplications; inversions and translocations and all combined). Overall, we found the heritability was better explained when combining the SNP data as well as the SVs data. In 45 traits SVs explained 25% or more of the trait variability. Five traits that were explained by over 90% heritability using SNPs and SVs came from different growth conditions in liquid medium. This may highlight again the influence of environmental conditions on the genomic structure. For 74 traits (~30% of those we analyzed) SVs explain more of the trait than the SNPs. These high SV-affected traits include malic acid, acetic acid and glucose/fructose contents of wine, key components of taste.
A collaborative effort
On a personal note, the paper concludes a wonderful team effort over two and a half years.
The project started as a summer project for Clemency Jolly, who had then just completed her 3rd undergraduate year at UCL, in the Dessimoz and Bähler labs. Dan Jeffares and the rest of the Bähler lab had just published their 161 fission yeast genomes, with an in-depth analysis of the association between SNPs and quantitative traits (Jeffares et al., Nature Genetics 2015). Studying SVs was the logical next step, but given the challenging nature of reliable SV calling, we also recruited to the team Fritz Sedlazeck, collaborator and expert in tool development for NGS data analysis then based in Mike Schatz’s lab at Cold Spring Harbor Laboratory.
At the end of the summer, it was clear that we were onto something, but there was still a lot be done. Clemency turned the work into her Master’s project, with Dan and Fritz redoubling their efforts until Clemency graduation in summer 2015. It took another year of intense work lead by Dan and Fritz to verify the calls, perform the GWAS and heritability analyses, and publish the work. Since then, Clemency has started her PhD at the Crick Institute, Fritz has moved to John Hopkins University, and Dan has started his own lab at the University of York.
References:
Jeffares, D., Jolly, C., Hoti, M., Speed, D., Shaw, L., Rallis, C., Balloux, F., Dessimoz, C., Bähler, J., & Sedlazeck, F. (2017). Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast Nature Communications, 8 DOI: 10.1038/ncomms14061
Carr AM, MacNeill SA, Hayles J, & Nurse P (1989). Molecular cloning and sequence analysis of mutant alleles of the fission yeast cdc2 protein kinase gene: implications for cdc2+ protein structure and function. Molecular & general genetics : MGG, 218 (1), 41-9 PMID: 2674650
Jeffares, D., Rallis, C., Rieux, A., Speed, D., Převorovský, M., Mourier, T., Marsellach, F., Iqbal, Z., Lau, W., Cheng, T., Pracana, R., Mülleder, M., Lawson, J., Chessel, A., Bala, S., Hellenthal, G., O’Fallon, B., Keane, T., Simpson, J., Bischof, L., Tomiczek, B., Bitton, D., Sideri, T., Codlin, S., Hellberg, J., van Trigt, L., Jeffery, L., Li, J., Atkinson, S., Thodberg, M., Febrer, M., McLay, K., Drou, N., Brown, W., Hayles, J., Salas, R., Ralser, M., Maniatis, N., Balding, D., Balloux, F., Durbin, R., & Bähler, J. (2015). The genomic and phenotypic diversity of Schizosaccharomyces pombe Nature Genetics, 47 (3), 235-241 DOI: 10.1038/ng.3215
Benito, A., Jeffares, D., Palomero, F., Calderón, F., Bai, F., Bähler, J., & Benito, S. (2016). Selected Schizosaccharomyces pombe Strains Have Characteristics That Are Beneficial for Winemaking PLOS ONE, 11 (3) DOI: 10.1371/journal.pone.0151102
More info
•
Author: Natasha Glover •
∞
Our paper “A Pragmatic Approach to Getting Published: 35 Tips for Early Career Researchers” just came out in Frontiers in Plant Science. This is the story behind the paper.
For my second postdoc, I was the fortunate receipient of a PLANT FELLOWS scholarship. PLANT FELLOWS is an international program that provides research grants to postdocs in the field of plant science. The fellows are based at many different host institutions throughout Europe. I myself am working at Bayer Crop Science in Gent, Belgium, in collaboration with the Dessimoz lab in London and Lausanne. Part of the PLANT FELLOWS mission is to provide training, mentoring, and networking to the postdocs—skills essential for career advancement.
Last year, the annual PF meeting was held in Männedorf, Switzerland from September 28 to October 1 2015. Training workshops took place at the Boldern Hotel, surrounded by meadows and with a nice view of Lake Zürich.

Group picture from the 3rd annual PLANT FELLOWS meeting
The meeting consisted of several days of trainings and workshops. For one of the days, I chose to participate in the workshop “Advanced Strategies for Dealing with the Publication Process.” I was especially keen on learning more about this particular subject. As a postdoc still trying to navigate the publication waters, I was looking for all the advice I could get. We’ve all heard the saying before: publish or perish. Publishing papers in your postdoc years is so important for an academic career.
There were about 15 postdocs in this day-long workshop. The facilitator, Philipp Mayer, came with a bunch of photocopied book chapters, articles, and USB keys full of pdfs for each of us to use on our laptops. The objective of the workshop was to, as a group, write a small paper about advanced publication strategies using the literature we were provided with. Our plan of attack was to pool our collective postdoc experience and come up with a list of our most useful recommendations on how to get a scientific paper published.
After feverishly reading websites, book chapters and papers, at the end of the day we came up with a draft: an introduction, our recommendations broken into 3 main sections, and a conclusion. We had a respectable number of references. But what would be the fate of our paper? About a third of the class was apathetic, a third thought we should aim for a blog post, and another third thought we should try for a “real” scientific journal. I had really enjoyed the workshop so I lobbied for publishing it in a real journal. I liked the experience of learning about a topic, working collaboratively with my peers, and then passing on the information for others to benefit.
I volunteered to take charge of the paper, edit it, and submit it to journals in hopes of getting it published. At the end of the day I left with a draft of the paper, many references, the contact information of all the attendees, and the full support of the facilitator (Philipp) for any future help that I might need. I looked at it as an opportunity take a leadership role in publishing a paper, from start to finish. And more importantly, it was a chance to put our own advice into practice.
Upon returning to Belgium, I quickly found out that one of the sentences we had written in the paper rang true: It is a common misconception among early career researchers that the presentation of the work in a manuscript is the last stage of a project. There is a long and complicated process associated with submission, review, and revision that must be taken into account. During the next month, I reread paper, finished writing short sections, added references, edited, and got feedback from the coauthors. We agreed on the author order, and shared the document using Authorea. Philipp and I went back and forth with several rounds of editing.
Attempt #1
We decided to submit our manuscript to eLife, which is a prestigious peer reviewed open access journal with favorable policy toward early career researchers. I wrote a cover letter to the editor describing our paper and asking if the topic was suitable to be considered for eLife.
Within a few days, the editor read the manuscript but informed me that he was unable to send it out for review because it wasn’t “fresh” enough, meaning most of what we said had already be discussed many times in the scientific community. Despite the sting of having a paper rejected directly from the editor, I decided to take the advice we had written in the paper: Remove your personal feelings from the peer review process. Time to find the next journal.
During the following month and a half, the manuscript was pushed to the bottom of my To Do list, as other projects and tasks got my attention. Christmas holidays came and went, and admittedly this paper was the last thing on my mind.
Attempt #2
In January, I sent a presubmission inquiry to PLOS Biology. The PLOS Biology editor wrote back within a few days to inform me that although they appreciated the attention to an important problem, they could not encourage us to submit because it didn’t present “novel strategies for increasing access to research, improving the quality of research results, or fixing flawed measures of impact.” Since this was the second time I had heard this same exact criticism, I realized it was time to take more advice from the paper: It is critical to highlight the novelty and importance in the article and cover letter. We were going to have to add something to the paper to make it more novel.
Attempt #3
Shortly after, I contacted the Frontiers in Plant Science (FiPS) Editorial Office with a new and improved cover letter. FiPS is an open access online journal publishing many different peer reviewed articles: research, reviews, commentaries, and perspectives, among others. The editor and I discussed morphing the paper into something that would be more plant related, given the plant science background of all the coauthors. Over the next month, it was back to editing the paper. I proposed edits that would make our tips more plant-specific. We added advice about industry-academia collaborations, and more information about plant science journals. Philipp, the coauthors, and I went back and forth several times with rounds of edits, adding more references and polishing more details. I submitted the final version of the paper to Frontiers in Plant Science on March 15.
The experience of the collaborative peer review by FiPS was a pleasant and efficient one. Their website says “Frontiers reviews are standardized, rigorous, fair, constructive, efficient and transparent.” I enthusiastically agree. Within two weeks, we had received comments from the reviewers. There were some major points that needed to be addressed before Frontiers could offer publication. However, the points were all very relevant and only helped to make the paper stronger. During the process of the interactive review, I took more guidance from the paper: Go point by point through the reviewer comments and either make the suggested change or politely explain and clarify the misunderstanding.
April 21st : Acceptance achieved! Approximately 5 weeks after submitting the article, it was accepted and the provisional version of the manuscript was published online. This is an extremely fast turnover time, in part due to the responsiveness of the editor, quick but in-depth peer review, and the interactive, transparent review discussion.
What I learned
This collaboration with the PLANT FELLOWS postdocs resulted in a paper I can say I’m proud of. I learned many things about the publication process—not only through a literature review, but by actually experiencing the process first hand. Here are some of the main things that stuck with me:
- There is a certain creative power in bringing people together in a beautiful location to brainstorm and produce an outcome within a short period of time. However, it is necessary for someone to take the reins and commit to the follow-through in order to get to a finished product. I think things like hackathons or other collaborative group efforts could lead to fruitful outcomes.
- I learned how to coordinate a small project. This was a great collaborative effort, which gave me an opportunity to practice the recommendations we wrote about in the paper.
I discovered firsthand the importance of the initial contact with the editor. As soon as we reworked the paper to approach the topic from a plant-specific standpoint, this added novelty to the paper. We were able to highlight this novelty in the cover letter.
- Don’t give up. Many times I got distracted or discouraged and thought to publish the manuscript on our blog, but I’m glad in the end we found a home for it at FiPS. Perseverance is key.
References
Glover, N., Antoniadi, I., George, G., Götzenberger, L., Gutzat, R., Koorem, K., Liancourt, P., Rutowicz, K., Saharan, K., You, W., & Mayer, P. (2016). A Pragmatic Approach to Getting Published: 35 Tips for Early Career Researchers Frontiers in Plant Science, 7 DOI: 10.3389/fpls.2016.00610
•
Author: David Dylus •
∞
The paper introducing our new tree visualisation tool Phylo.io was just published in MBE.
Yet another tool to display trees, you might say, and indeed, so it is. But for all the tools that have been developed over the years, there are very few that scale to large trees, make it easy to compare trees side-by-side, and simply run in a browser on any computer or mobile device.
To fill this gap, we created Phylo.io.
Story behind the paper
The project started as a student summer internship project, with the aim of producing a tree visualiser that facilitates comparison of trees built on the same set of leaves. After reading the project description, Oscar Robinson, a brilliant student from the Computer Science department at UCL, decided to work on this project during a three month internship. He saw a chance to apply his experience in the development of web tools and to develop his knowledge in the field of data visualisation, one of his major interests.
Once Oscar started with the development of Phylo.io, he realised that only a few tools existed for visual comparison of two trees and either seemed to rely on old technology or were cumbersome to use. Especially this incentive lead him to develop our tool into a fully fledged online resource that is easy to use, platform independent and based on the newest javascript libraries (e.g. D3). Within three months, he managed to produce a prototype of the tool. However, due to the short length of the internship, some details still needed a bit of attention.
Luckily for me, I started my PostDoc in the Dessimoz Lab around that time. Being a novice in a computational lab, Christophe proposed to me to take over the project and bring it to completing as a way to kickstart my postdoc. Altough my computational background at that time did not include any experience in JavaScript programming, I anyway accepted the challenge and was eager to start learning the material. Especially my initial steep learning progress was facilitaed by the help of two other brilliant PhD students, Alex Vesztrocy and Clément Train. Once I acquired some basic understanding, I was able to resolve bugs and add some key missing functionalities such as automatic tree rerooting or persistent storage and sharing functionality.
What is phylo.io and what can it do?
Phylo.io is a web tool that works in any modern browser. All computations are performed client-side and the only restriction on performace is the machine it is running on. Trees can be input in Newick and Extended Newick format. Phylo.io offers many features that other tree viewers have. Branches can be swapped, the rooting can be changed, the thickness, font and other parameters are adaptable. Many of these operations can be performed directly by clicking on a branch or a node in the tree. Importantly, it features an automatic subtree collapsing function: this facilitates the visualisation of large trees and hence the analysis of splits that are deep in the tree.
Next to basic tree visualisation/manipulation it features a compare mode. This mode allows to compare two trees computed using different tools or different models. Similarities and differences are highlighted using a colour scheme directly on the individual branches, making it clear where the differences in two topologies actually are. Additionally, since the output of different tools provides trees with very different rootings and leaf order, Phylo.io has a function to root one of the trees according to the other one and adapt the order of the leaves according to a fixed tree.
How do you use phylo.io?
To save you time, here is a one minute screencast highlighting some of the key features of Phylo.io:
You can find more info in the Phylo.io Manual.
Reference
Robinson, O., Dylus, D., & Dessimoz, C. (2016). Phylo.io: interactive viewing and comparison of large phylogenetic trees on the web
Molecular Biology and Evolution DOI: 10.1093/molbev/msw080
 The Dessimoz Lab
blog is licensed under a Creative
Commons
Attribution 4.0 International License.
The Dessimoz Lab
blog is licensed under a Creative
Commons
Attribution 4.0 International License.